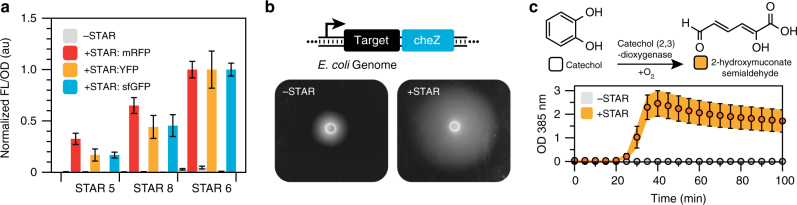
Fig. 3.
STARs control gene expression tightly in diverse contexts. a STARs function in the context of sequence diverse fluorescent reporter genes. Fluorescence characterization was performed on E. coli cells transformed separately with plasmids encoding three target RNAs controlling sfGFP, YFP or mRFP in the absence (-STAR) and presence (+STAR) of plasmids encoding cognate STARs. Measurements were normalized to 1 for the fluorescence value of STAR variant 6 in the +STAR condition for each fluorescent protein. Fluorescence characterization was performed by bulk fluorescence measurements (measured in units of fluorescence [FL]/optical density [OD] at 600 nm). b STARs function in the genome to control endogenous E. coli genes. Schematic of STAR regulated cheZ construct integrated into E. coli BW25113ΔcheZ. Photographs of semi-solid agar motility assays in the absence (left panel, -STAR) and presence (right panel, +STAR) of a plasmid encoding the cognate STAR. Motility is indicated by the formation of halos around the inoculation site in the center of the photographs. c STARs function in cell-free transcription-translation (TX-TL) reactions to tightly control enzyme expression. Schematic of the catechol (2,3)-dioxygenase catalyzed reaction that converts colorless catechol into a yellow-colored 2-hydroxymuconate semialdehyde. Spectral characterization of observed yellow color (measured in units of OD at 385 nm) of STAR controlled catechol (2,3)-dioxygenase expression in TX-TL reactions supplemented with catechol in the absence (-STAR) and presence (+STAR) of a plasmid encoding cognate STAR. Data in a, c represent mean values and error bars represent s.d. of n = 9 biological replicates, and b is a representative photograph of n = 1 biological replicates with repeats shown in Supplementary Fig. 16