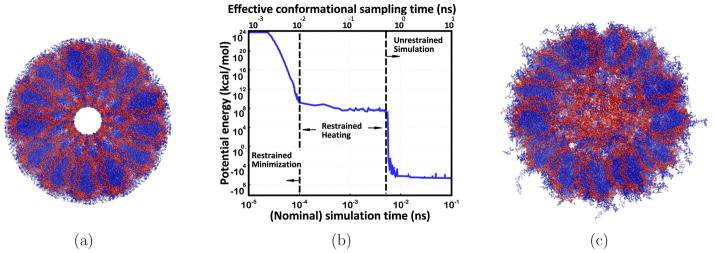
Figure 4.
(a) The manually constructed model13 of ~ 1.16 million atom chromatin fiber (consistent with low resolution cryo-EM data86) is energetically unrealistic; no atomically detailed experimental structures are available. (b) A 0.1 ns GB-HCPO simulation of the fiber significantly reduces the steric clashes, as seen by the large reduction in the potential energy. Data points represent averages over 100 time step intervals (1 fs each). In terms of sampling the conformational transitions involved, the 100 ps simulation performed here is effectively equivalent to ~10 ns explicit-solvent TIP3P PME simulation (the conformational sampling is ~100 folds faster66), denoted as effective conformational sampling time. (c) Equilibrated structure (all-atom MD, GB-HCPO) suggests important structural details consistent with experimental results: the linker DNA fills the core region, the H3 histone tails interact with the linker DNA.107,108