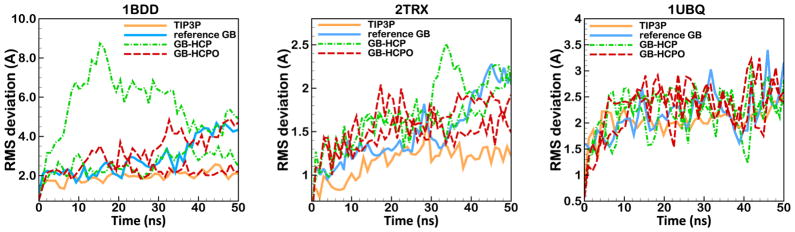
Figure 9.
Backbone RMS deviation from the experimental reference structure for 50 ns MD simulation based on GB-HCPO (red dashed line) compared to the reference explicit-solvent method (TIP3P), reference GB (without approximation), and GB-HCP. The test structures are, from left to right: immunoglobulin binding domain (1BDD, residues 10–55), thioredoxin (2TRX, all residues), and Ubiquitin (1UBQ, all residues). Two independent trajectories were produced for each of GB-HCPO and GB-HCP approximations. Each trajectory is sampled every 1 ns. Connecting lines are shown to guide the eye.