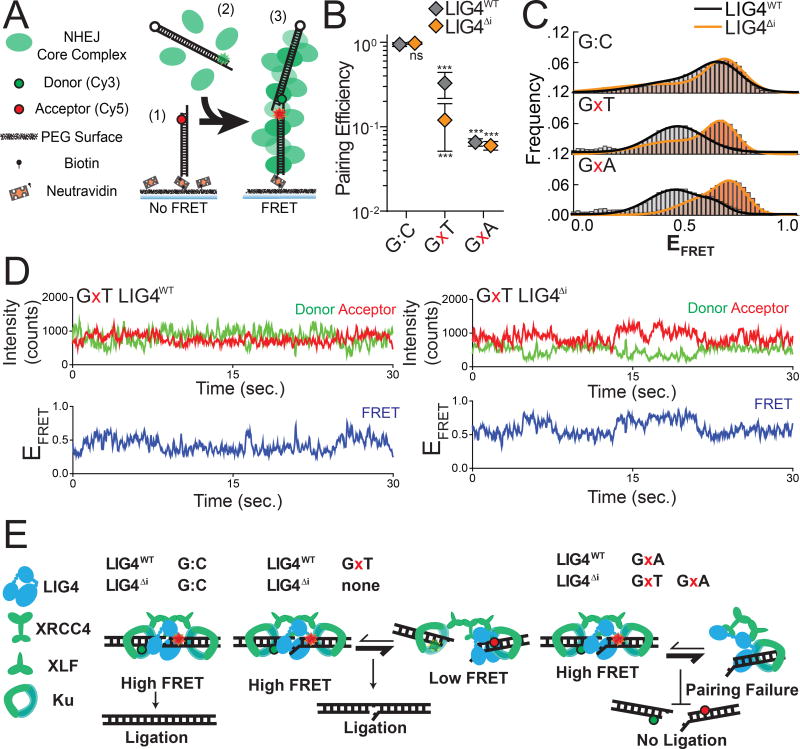
Figure 2.
Effect of complex end structures on pairing dynamics of single molecule complexes with LIG4WT or LIG4ΔI (A) smFRET NHEJ assay: (1) dsDNA with a Cy5 acceptor is tethered to a bitonylated PEG surface via a biotin-neutravadin linkage, (2) dsDNA with a Cy3 donor and NHEJ proteins (green) are added to the chamber, and (3) ends are paired and FRET is observed. (B) Quantitation of pairing efficiency of ends with complementary (G:C) or mismatched (GxT, GxA) overhangs by Ku, XLF, XRCC4 and either LIG4WT (gray) or LIG4ΔI (orange). Error bars represent standard error of the mean for 3 experiments. Means were assessed by two-way ANOVA as significantly different from control (LIG4WT on G:C substrate) with confidence p<0.001 (***). (C) Histograms of observed EFRET for PECs formed as in (B). (D) Representative smFRET trajectory for LIG4WT and LIG4ΔI PECs formed with GxT ends demonstrating altered transition frequency and FRET states (E) LIG4WT enables PECs to oscillate between high and low EFRET states in response to distortions, and this flexibility is essential for joining distorted breaks.