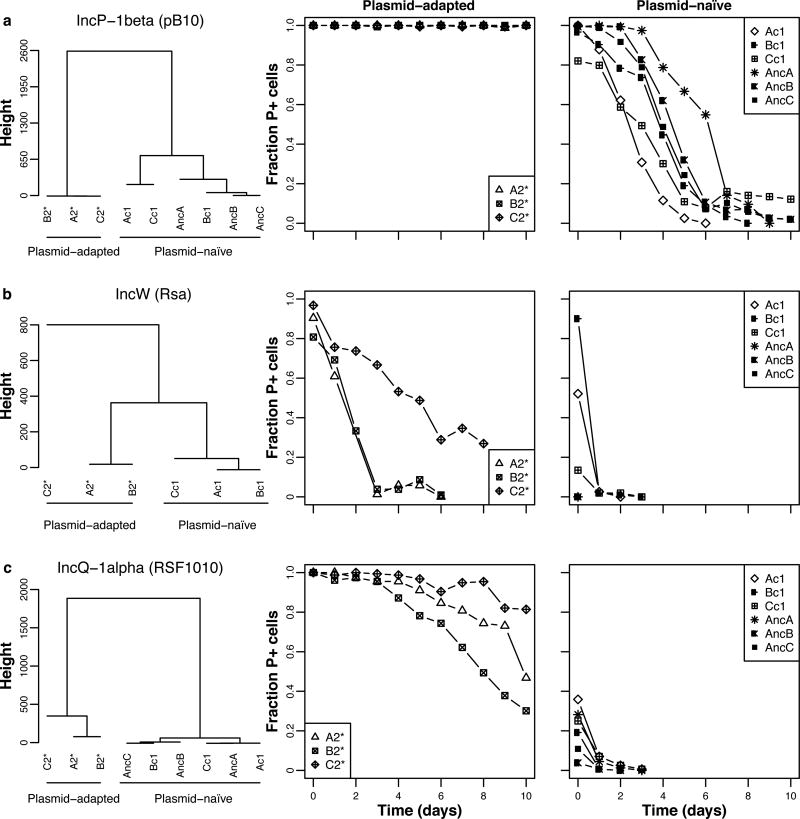
Figure 4. Pseudomonas sp. nov. H2 evolved to be more permissive towards both related and unrelated plasmids.
The plasmid persistence profiles in three clones each of RP4-adapted, ancestral and control hosts are shown for three naive plasmids: (A) the closely related IncP-1 β plasmid pB10, (B) the distantly related IncW plasmid Rsa, and (C) the unrelated IncQ-1α plasmid RSF1010. They always clustered into two distinct groups: Group A contained the three RP4-adapted hosts and Group B the ancestral and control hosts, naïve to the plasmid. Note that due to the extremely low persistence of plasmids Rsa and RSF1010, the initial fractions of plasmid-containing (P+) ancestral cells were not close to 1, even though the pre-culture contained plasmid-selective antibiotics. This is due to survival of plasmid-free cells in the pre-cultures and additional plasmid loss in the colonies during replica-plating. Moreover, the persistence of Rsa in the ancestral host was so poor that no temporal data could be collected to compare to the other Rsa persistence profiles. Each data point represents the mean fraction of P+ cells (n = 3). See also Fig. S3 and Table S4 for the modeled predictions.