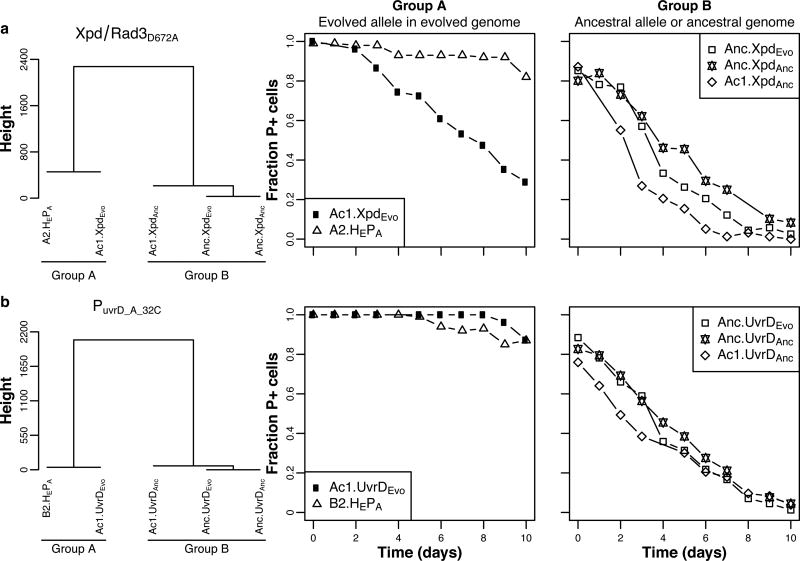
Figure 6. At least two mutations were required for full plasmid persistence, one plasmid-adaptive and one environment-adaptive mutation.
Complete linkage cluster analysis (left) of the persistence profiles (shown to the right) of plasmid-adapted clones A2 (A2.HEPA in panel A) and B2 (B2.HEPA in panel B), and control clone Ac1 and ancestral clone Anc, each containing either the evolved or ancestral allele of Xpd/Rad3D672A (denoted as XpdEvo or XpdAnc) (A), and PuvrD_A-32C (UvrDEvo or UvrDanc) (B). See also Supplemental Fig. 7 and Supplemental Tables 6, 7, 8 for the modeled predictions. Each data point represents the mean fraction of plasmid-containing cells (n = 3).