Figure 2.
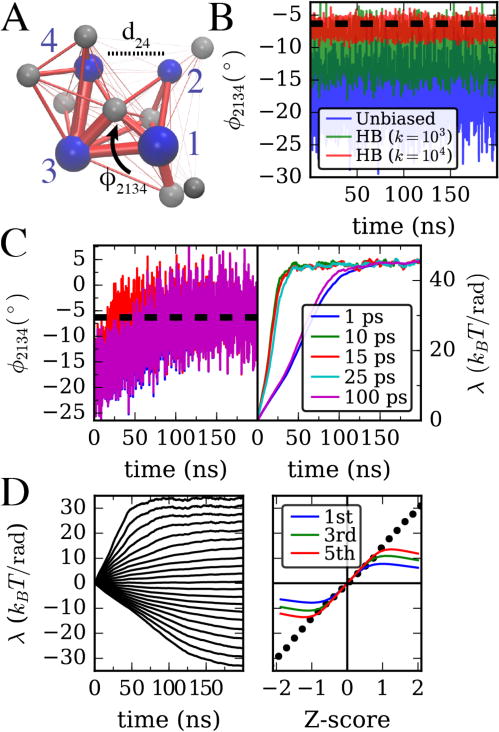
(A) Twelve-site hENM of an ATP-bound actin monomer parameterized as described in the main text. The four major subdomains of actin are labeled, and cleft distance and twist angle CVs are defined as in Figure 1A. (B) Twist angle for an unbiased hENM, as well as with a harmonic bias with force constants 103 and 104 kJ/mol/rad2 centered at (dashed line). (C) Twist angle evolution as well as biasing parameter using gradient descent algorithm of Ref. 8 is shown for different τavg. (D) Left, bias parameter as in C (τavg = 10ps) with target values for ϕ from −25.2° to −9.16°. Right, comparison of final bias parameters on left (dots) with first, third, and fifth order predictions given in the main text. The horizontal axis shows difference of target ϕ from ϕunbiased scaled by the unbiased standard deviation as computed from data in (B).
