Abstract
Serological analysis of recombinant tumor cDNA expression library (SEREX) is a powerful and widely used method to explore the cancer immune environment. In the present study, immunoscreening of normal testicular tissues and malignant mesothelioma (MM) cancer MSTO-211H cell line cDNA libraries with sera from 5 MM patients led to the isolation of 16 independent antigens, which were designated ‘Korea Pusan-Malignant Mesothelioma’ (KP-MM)-1 to −16. In total, 3/16 antigens were identified using the results of previous SEREX analyses, and 13 were newly identified. Of these, KP-MM-8, which was subsequently identified as amyotrophic lateral sclerosis 2 chromosome region candidate 11, was shown to be tissue-restricted. Reverse transcription-polymerase chain reaction demonstrated KP-MM-8 to be expressed strongly only in the normal testis, and weakly in the spleen, prostate, ovary, heart and skeletal muscle. In addition, KP-MM-8 mRNA was identified in MM cell lines, and in various other cancer cell lines, including MM (3/4), lung cancer (5/7), melanoma (5/7) and liver cancer (5/5) cell lines. Additionally, 2/16 antigens (KP-MM-2 and KP-MM-6) exclusively reacted with sera from cancer patients. However, KP-MM-8 reacted with 1 of 8 MM sera. Notably, 8/8 patients with MM and 8/8 normal individuals exhibited antibodies reactive to KP-MM-5, which was identified as cell division cycle 25B, a known oncogene. Overall, this data suggests that KP-MM-8 may be considered as a cancer/testis-like antigen and KP-MM-5 as an immunogenic tumor antigen in MM patients.
Keywords: SEREX, cancer/testis antigen, malignant mesothelioma, tumor antigen
Introduction
Malignant mesothelioma (MM) is a rare but rapidly fatal cancer that can develop from mesothelial cells in the pleura, peritoneum, pericardium and tunica vaginalis (1,2). The latency period of MM is exceptionally long and patients diagnosed with MM generally have poor prognoses (3), with an average life expectancy of less than a year, dependent on the tissue in which it originates (4,5). It has been reported that asbestos is the main carcinogen associated with MM (6,7); however, the molecular mechanism responsible for the pathogenesis of MM has not been elucidated.
Standard therapies for MM include chemotherapy, surgery and radiotherapy, but more effective strategies are required to improve the result of conventional therapeutic methods or cure this cancer. Immunotherapy may be a useful treatment (7,8); however, despite research progress, immunotherapy using tumor-associated antigens has been barely attempted in MM. Tumor antigens are classified into several categories, including differentiation antigens (9), mutated gene products (10), overexpressed oncogenes (11) and cancer/testis (CT) antigens (12,13). Of these, CT antigens are considered a promising tumor antigen group, since these proteins are expressed in the normal testis and tumor tissues (14,15).
Various methods have been devised to identify this group of tumor antigens. Serological analysis of recombinant tumor cDNA expression library (SEREX) is one such method, which has been shown to be powerful and effective (15). This technique screens cDNA expression libraries using the sera of cancer patients, and identifies cancer antigens recognized by autologous serum IgG (16). At present, >2,500 tumor antigens, including melanoma-associated antigen (MAGEA) (13), synaptonemal complex protein 1 (17), NY-ESO-1 (18), NY-SAR-35 (19) and KP-CoT-23 (20) have been identified using SEREX. Certain antigens, such as MAGE-A3 and NY-ESO-1, have been evaluated for immunogenicity and for their immunotherapeutic efficacies (21). In addition, CT antigens are expected to be utilized as biomarkers for diagnosis and prognosis (21,22).
In the present study, SEREX methodology was applied to define the spectrum of immunogenic proteins in MM, with focus on the CT-like antigens KP-MM-8 and KP-MM-5, which demonstrated high seroreactivity in patients with MM.
Materials and methods
Human sera and cell lines
Human sera from 8 healthy donors and 8 MM patients were obtained between February 2008 and December 2013, from the Department of Pathology and Environmental Health Center for Asbestos, Pusan National University Hospital (Yangsan, Korea) subsequent to diagnosis and staging. Human MM NCI-H226, NCI-H2452, NCI-H28 and MSTO-211H cell lines, human lung cancer A-427 and A549 cell lines, and the human melanoma cancer A375 cell line were obtained from the American Type Culture Collection (Manassas, VA, USA). Human colon cancer SNU-C1, SNU-C2A, SNU-C4 and SNU-C5 cell lines, human ovarian cancer SNU-8 and SNU-840 cell lines were obtained from the Korean Collection for Type Cultures (Jeongeup, Korea). All cell lines were maintained in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum medium (Gibco; Thermo Fisher Scientific, Inc.), 2 mM L-glutamine, 100 units/ml penicillin and 100 µg/ml streptomycin. Cells were cultured at 37°C in a humidified 5% CO2 atmosphere. The present study was performed with approval from the Ethical Committee of Pusan National University Yangsan Hospital (Yangsan, Korea).
Total RNA extraction from cell lines
Total RNA of human malignant mesothelioma NCI-H226, NCI-H2452, NCI-H28 and MSTO-211H cell lines, human lung cancer A-427 and A549 cell lines, human colon cancer SNU-C1, SNU-C2A, SNU-C4 and SNU-C5 cell lines and human ovarian cancer SNU-8 and SNU-840 cell lines were isolated using a RNA isolation kit (RNeasy Maxi kit; Qiagen, Hilden, Germany). Amounts of RNA isolated were assessed at 260 nm using a spectrophotometer (Ultrospec 2000; Pharmacia Biotech; GE Healthcare, Chicago, IL, USA). Total RNAs from: Human colon cancer HCT-15, SK-CO-1, SW-1116 and KM12/L4A cell lines; human lung cancer SK-LC-14, SK-LC-19, SK-LC-5, NCI-H23 and NCI-H740 cell lines; human melanoma cancer A375/C5, A375/P, A375/SM, SK-MEL-19, SK-MEL-128, SK-MEL-21 and SK-MEL-37 cell lines; human ovarian cancer OV-CAR-3, and SK-OV-3 cell lines; human breast cancer SK-OV-1, BT-474, MDA-MB-453, MCF-7, MDA-MB-231 and SK-BR-5 cell lines; human renal cancer SK-RC-18 and SK-RC-59 cell lines; human leukemia CEM/S and K-562 cell lines; human rhabdomyosarcoma A-204 cell lines; human thyroid cancer KAT-18 cell lines; and human liver cancer SK-HEP-1, SNU-354, SNU-423, SNU-449 and SNU-475 cell lines were obtained from the Ludwig Institute for Cancer Research (Memorial Sloan-Kettering Cancer Center, New York, NY, USA). Normal tissue total RNA was purchased from Clontech Laboratories, Inc. (Mountain View, CA, USA, cat. nos. 636742 and 636743).
Preparation of the cDNA library and sera
Poly(A)+ RNA from normal testis was purchased from Clontech Laboratories Inc. mRNA of the MSTO-211H cell line was extracted using the PolyATtract mRNA Isolation System (Promega Corp., Madison, WI, USA). Poly(A)+ RNA (5 µg) was used to construct a cDNA library in ZAP Express vector (Stratagene; Agilent Technologies, Inc., Santa Clara, CA, USA), according to the manufacturer's instructions. The cDNA library contained approximately one million recombinants and was used for immunoscreening without amplification.
Sera from 8 healthy individuals and 8 patients with MM were diluted with 0.2% skimmed milk to 1:200. To remove serum antibodies that react with Escherichia coli/bacteriophage-associated antigens, sera were absorbed with E. coli/bacteriophage lysates, as previously described (20).
Immunoscreening of the cDNA library
Immunoscreening of the cDNA library was performed as described by Song et al (20). Briefly, E. coli XL1 blue MRF cells (Stratagene; Agilent Technologies, Inc.) were transfected with the recombinant phages, plated at a density of ~5,000 pfu/150 mm plate (NZCYM-IPTG agar), and incubated for 8 h at 37°C. The cells were then transferred to nitrocellulose filters (PROTRAN BA 85; 0.45 µm; Schleicher & Schuell, Keene, NH, USA). The filters were incubated with patient sera, which had been preabsorbed with E. coli-phage lysate, at a dilution of 1:200, at room temperature overnight. The serum-reactive clones were detected using alkaline phosphatase-conjugated secondary antibody (Abcam, Cambridge, UK; cat. no., ab6859; dilution, 1:2,000) incubated for 1 h at room temperature and then visualized by incubation with 5-bromo-4-chloro-3-indolylphosphate/nitroblue tetrazolium. Subsequent to screening, isolated positive clones were removed from plates and preserved in suspension medium buffer with 25 µl chloroform.
Positive phages were mixed with a helper phage to co-infect XL-1 Blue MRF cells, and then rescued into pBluescript phagemid forms by in vivo excision. Excised phagemids were transformed into host bacteria (XLOLR) to multiply for plasmid extraction and stock. The size of the cDNA inserted was determined by double restriction enzyme digestion with EcoRI and XhoI. The cDNA was sequenced commercially (Macrogen, Inc., Seoul, Korea).
Reverse transcription-polymerase chain reaction (RT-PCR)
The cDNA preparations used as templates for RT-PCR reactions were prepared using 1 µg total RNA isolated from all cell lines and the Superscript First Strand Synthesis kit (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA). Primers were as follows: Amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 11 (ALS2CR11) forward, 5′-TCTGCAGGCTGGAGGTGGAA-3′ and reverse, 5′-CCTTTCCCGGGGTTGTTGCT-3′. The conditions used for RT-PCR were initial activation for 5 min at 94°C, 35 amplification cycles of denaturation for 30 sec at 94°C, and annealing for 30 sec at 63°C, extension for 1 min at 72°C, followed by a final elongation for 10 min at 72°C. A total of 20 µl of PCR product were loaded onto a 1.2% agarose gel and electrophoresis was performed at room temperature at 100 V 30 min using 1X Tris-acetate-EDTA buffer. The gels were stained with ethidium bromide solution diluted with distilled water to a ratio of 2:104 for 10 min, and washed with H2O. Data were analyzed by Gel Doc™ + Imager with Image Lab™ software v.5.2.1 (Bio-Rad, Richmond, CA, USA).
Seroreactivity by plaque assay
Selected immunoreactive clones were tested for reactivity against sera from individual MM patients and healthy donors using a plaque assay. λ-ZAP clone without an insert was co-plated and included in each assay as a negative control. KP-MM-5 was tested for reactivity against 10-fold serially diluted serum (1:103 to 1:106) using the same plaque assay.
Results
Identification of MM antigens by SEREX
To identify tumor antigens in MM, immunoscreening of a testis cDNA library and MSTO-211H cDNA library was performed using the sera of patients with MM. In total, 31 immunogenic clones were identified in this manner, and 16 of the 31 genes were verified as tumor antigen candidates and termed KP-MM-1 to KP-MM-16 (Table I). Of the 16, 3 were previously identified by SEREX in other tumor types, and 13 antigens did not match entries in the cancer immunome database, and were therefore considered new antigens.
Table I.
Malignant mesothelioma antigens identified by SEREX.
| Antigen | Gene name | UniGene clustera | Library source | Serum sourceb | Redundancy, n | Previously identified by SEREXc |
|---|---|---|---|---|---|---|
| KP-MM-1 | IFT74 | Hs.145402 | Testis | M4 | 1 | No |
| KP-MM-2 | TMF1 | Hs.267632 | Testis | M4 | 2 | Yes |
| KP-MM-3 | BRAP | Hs.530940 | Testis | M4 | 1 | Yes |
| KP-MM-4 | RPLP0 | Hs.546285 | MSTO-211H | M8 | 1 | Yes |
| KP-MM-5 | CDC25B | Hs.153752 | Testis | M8 | 2 | No |
| KP-MM-6 | ZNF429 | Hs.572567 | Testis | M49 | 1 | No |
| KP-MM-7 | ZNF320 | Hs.369632 | Testis | M49 | 1 | No |
| KP-MM-8 | ALS2CR11 | Hs.335788 | Testis | M8 | 1 | No |
| KP-MM-9 | DNAJC2 | Hs.558476 | Testis | M8 | 1 | No |
| KP-MM-10 | NUB1 | Hs.647082 | Testis | M10 | 1 | No |
| KP-MM-11 | CCDC11 | Hs.658630 | Testis | M1 | 2 | No |
| KP-MM-12 | ZNF93 | Hs.723768 | Testis | M49 | 1 | No |
| KP-MM-13 | ZNF234 | Hs.235992 | Testis | M49 | 1 | No |
| KP-MM-14 | ZNF595 | Hs.709469 | Testis | M49 | 1 | No |
| KP-MM-15 | CCDC40 | Hs.202542 | Testis | M49 | 1 | No |
| KP-MM-16 | ZNF85 | Hs.37138 | Testis | M49 | 1 | No |
UniGene cluster of isolated antigens (http://www.ncbi.nim.nih.gov/).
Sera from malignant mesothelioma patients: M1, a patient diagnosed with malignant mesothelioma; M4, a patient diagnosed with malignant peritoneal mesothelioma; M8, a patient diagnosed with malignant pleural mesothelioma; M10, a patient diagnosed with malignant pleural mesothelioma; and M49, a patient diagnosed with malignant peritoneal mesothelioma.
Sequences were compared with those in the SEREX database of the Ludwig Institute for Cancer Research (http://ludwigsun5.unil.ch/CancerImmuno meDB/). ‘Yes’ and ‘No’ indicate whether the antigen matched or did not match an antigen in the SEREX database, respectively.
KP-MM-8 as a CT-like antigen
Preliminary in silico mRNA expression profile analysis and characterization of the gene products identified in the present study were undertaken based on the tissue distributions of expressed sequence tags and serial analysis of gene expression tags in the Cancer Genome Anatomy Database (http://cgap.nci.nih.gov/) and the information contained in the GeneCards database (http://www.genecards.org/). One antigen, KP-MM-8, was identified as a CT-like antigen. KP-MM-8 was identified as ALS2CR11 by a nucleotide BLAST search.
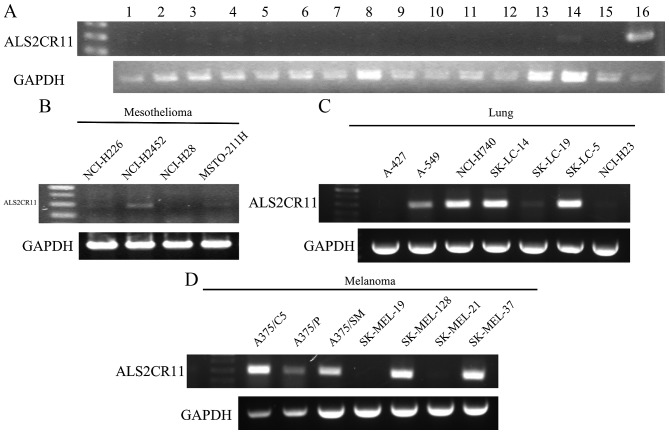
Conventional RT-PCR was performed using mRNA from normal tissues to examine the expression of ALS2CR11 (Fig. 1A). RT-PCR showed that ALS2CR11 was strongly expressed only in the normal testis, and weakly in the spleen, prostate, ovary, heart and skeletal muscle. However, the expression in various cancer cell lines was relatively high. The expression of this antigen was positive in 3 of 4 MM cell lines, consisting of the NCI-H226, NCI-H2452 and MSTO-211H cell lines (Fig. 1B). In addition, it was expressed in various cancer cell lines, including lung cancer (5/7) (Fig. 1C), melanoma (5/7) (Fig. 1D) and liver cancer (5/5) cell lines, but was not expressed in colon cancer, renal cancer or leukemia cell lines (Table II). This result suggests that ALS2CR11 may be a putative CT like antigen.
Figure 1.
Reverse transcription-polymerase chain reaction analysis of ALS2CR11 expression in (A) normal tissues, as follows: 1, spleen; 2, thymus; 3, prostate; 4, ovary; 5, small intestine; 6, colon; 7, leukocyte; 8, heart; 9, brain; 10, placenta; 11, lung; 12, pancreas; 13, liver; 14, skeletal muscle; 15, kidney; and 16, testis. ALSC2CR11 expression is also shown in (B) mesothelioma cancer, (C) lung cancer and (D) melanoma cell lines. cDNA templates were normalized using GAPDH as shown in the bottom panel. ALS2CR11, amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 11.
Table II.
Summary of ALS2CR11 mRNA expression as determined by reverse transcription-polymerase chain reaction.
| Cancer cell line | Frequency, n |
|---|---|
| Mesothelioma | 3/4 |
| Colon cancer | 0/8 |
| Lung cancer | 5/7 |
| Melanoma | 5/7 |
| Ovarian cancer | 2/4 |
| Breast cancer | 1/6 |
| Renal cancer | 0/2 |
| Leukemia | 0/2 |
| Rhabdomyosarcoma | 1/1 |
| Thyroid cancer | 1/1 |
| Liver cancer | 5/5 |
ALS2CR11, amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 11.
Seroreactivity of isolated mesothelioma antigens
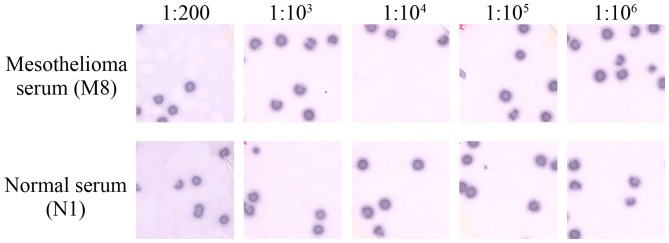
Seroreactivity testing of 7 phage clones, consisting of KP-MM-2, KP-MM-5, KP-MM-6, KP-MM-7, KP-MM-8, KP-MM-9 and KP-MM-12, was performed using the sera of 8 MM patients and 8 healthy donors by phage plaque assay (Fig. 2). The CT-like antigen KP-MM-8 was only positive in one MM serum (M8), and negative in all other 7 MM sera and to all normal sera. In total, 2 of the 8 MM patients had antibodies reactive to KP-MM-6-containing phagemids, whereas the 8 normal sera showed no reactivity against KP-MM-6 (Table III). Notably, all MM patients and normal individuals showed high reactivity to the KP-MM-5 antigen. Representative phage plaque assay for KP-MM-5 from the sera of M8 and normal individual N1, showed high-titer reactivity (1:106) (Fig. 3). The presence of consistently high-titer antibodies against KP-MM-5 suggested it to be an immunogenic tumor antigen in MM patients. However, normal sera also showed extremely strong reactivity for KP-MM-5; this observation requires further investigation.
Figure 2.
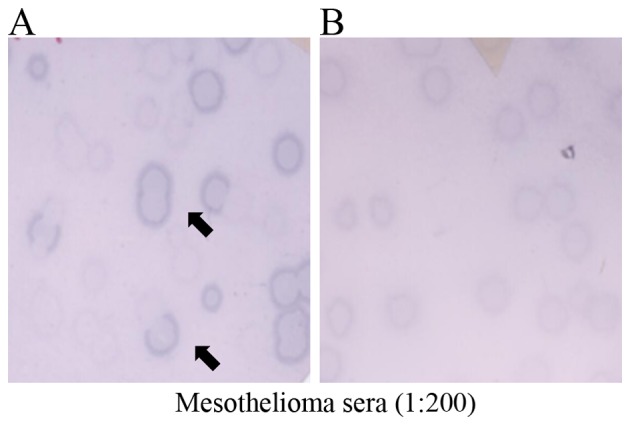
Representative phage plaque assays of the sera of the mesothelioma patients (A) M49 and (B) M11 for KP-MM-7. λ-ZAP phages without insert were mixed with KP-MM-7 clones and acted as internal negative controls. Mixtures were transfected into E. coli, and plaques were blotted onto nitrocellulose membranes and incubated with the sera of mesothelioma patients (dilution, 1:200). Spots were evaluated as (A) positive when clones were clearly distinguishable from control phage and as (B) negative if not. Arrows indicate positive clones.
Table III.
Detection of selected antigens against sera obtained from malignant mesothelioma patients and normal individuals.
| No. of cases | ||
|---|---|---|
| KP-MM-antigen | Mesothelioma | Normal |
| KP-MM-2 | 3/8 | 0/8 |
| KP-MM-5 | 8/8 | 8/8 |
| KP-MM-6 | 2/8 | 0/8 |
| KP-MM-7 | 1/8 | 0/8 |
| KP-MM-8 | 1/8 | 0/8 |
| KP-MM-9 | 2/8 | 2/8 |
| KP-MM-12 | 1/8 | 0/8 |
Figure 3.
Seroreactivity of the malignant mesothelioma patient M8 and the normal healthy individual N1 for KP-MM-5 (CDC25B). Serum dilutions are indicated. λ-ZAP phages without an insert were mixed with test clones and acted as internal negative controls; these are visible as a background to positive clones. Assays were scored positive only if test clones were clearly distinguishable from control phages.
Discussion
Previously, it was reported that several major CT antigens are expressed in MM (23); however, it is theorized that numerous tumor antigens remain unidentified, and that even identified tumor antigens require further functional study. To identify new CT antigens, MM was studied using the SEREX technique, and as a result, 31 sequences were isolated and 16 antigens were identified as independent tumor-specific antigens.
Notably, 6 of the 16 identified genes (KP-MM-6, KP-MM-7, KP-MM-12, KP-MM-13, KP-MM-14 and KP-MM-19) belonged to the zinc finger gene family. Associations between the ZNF gene and cancer properties are not well understood, although it has been reported that certain ZNF genes have potential as biological markers of cancer (24). Thus, additional investigation of the functional roles of ZNF gene during the pathogeneses of cancers, including MM, is required.
KP-MM-8 was identified in a testis library that reacted to mesothelioma serum, and BLAST search identified this gene as ALS2CR11, a member of the ALS2CR multigene family, which is associated with amyotrophic lateral sclerosis (25). Nevertheless, the present study identifies the ALS2CR11 gene as putative CT antigen, and little is known about the contribution made by this gene to carcinogenesis.
In the present study, ALS2CR11 was found to have CT-like antigen features, particularly a restricted mRNA expression pattern. Considering the mRNA expression profile of ALS2CR11 in the GeneCards database and the present RT-PCR results, ALS2CR11 appears to be expressed in various cancer cell lines and in few normal tissues, which is a noteworthy feature of CT antigens.
However, seroreactivity testing showed KP-MM-8 reacted with only 1 MM serum, although this result was based on a limited number of patient sera samples. Additional investigation is required with more samples to determine whether ALS2CR11 functions as a CT antigen, and to identify the molecular basis for its contribution to tumorigenesis.
KP-MM-5 was identified in a testis library that reacted with mesothelioma serum. This clone was identified as cell division cycle (CDC) 25B, and CDC25 phosphatases activate cyclin-dependent kinases (CDKs) and targets of the checkpoint kinase (CHK) 1/CHK2-mediated checkpoint pathway (26). Numerous studies have been conducted on members of the CDC25 protein phosphatase family, and it has been reported that CDC25B plays an important role in tumorigenesis (27,28). Specifically, CDC25B activates M phase promoting factor and promotes G2/M transition, and the overexpression of CDC25B contributes to excessive cell division (29).
Presence of high-titer reactive antibodies against KP-MM-5 suggests that KP-MM-5 is an immunogenic tumor antigen in MM patients. However, normal sera showed strong reactivity for KP-MM-5. This result requires additional investigation, and suggests that numerous aspects of the actions of CDC25B have yet to be identified.
Acknowledgements
The study was supported by a grant from Pusan National University Yangsan Hospital Environmental Health Center for Asbestos (Korea Ministry of Environment, grant no. 076-1900-1939-302-320-01).
References
- 1.Raptopoulos V. Peritoneal mesothelioma. Crit Rev Diagn Imaging. 1985;24:293–328. [PubMed] [Google Scholar]
- 2.Cunha P, Luz Z, Seves I, Sousa C, Skiappa, Ribeiro L, Marques C, Oliveira M. Malignant peritoneal mesothelioma-diagnostic and therapeutic difficulties. Acta Med Port. 2002;15:383–386. (In Portuguese) [PubMed] [Google Scholar]
- 3.Ahmed I, Tipu Ahmed S, Ishtiaq S. Malignant mesothelioma. Pak J Med Sci. 2013;29:1433–1438. doi: 10.12669/pjms.296.3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Røe OD, Stella GM. Malignant pleural mesothelioma: History, controversy and future of a manmade epidemic. Eur Respir Rev. 2015;24:115–131. doi: 10.1183/09059180.00007014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mirabelli D, Roberti S, Gangemi M, Rosato R, Ricceri F, Merler E, Gennaro V, Mangone L, Gorini G, Pascucci C, et al. Survival of peritoneal malignant mesothelioma in Italy: A population-based study. Int J Cancer. 2009;124:194–200. doi: 10.1002/ijc.23866. [DOI] [PubMed] [Google Scholar]
- 6.Rudd RM. Malignant mesothelioma. Br Med Bull. 2010;93:105–123. doi: 10.1093/bmb/ldp047. [DOI] [PubMed] [Google Scholar]
- 7.Robinson BW, Lake RA. Advances in malignant mesothelioma. N Engl J Med. 2005;353:1591–1603. doi: 10.1056/NEJMra050152. [DOI] [PubMed] [Google Scholar]
- 8.Kondola S, Manners D, Nowak AK. Malignant pleural mesothelioma: An update on diagnosis and treatment options. Ther Adv Respir Dis. 2016;10:275–288. doi: 10.1177/1753465816628800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coulie PG, Brichard V, Van Pel A, Wölfel T, Schneider J, Traversari C, Mattei S, De Plaen E, Lurquin C, Szikora JP, et al. A new gene coding for a differentiation antigen recognized by autologous cytolytic T lymphocytes on HLA-A2 melanomas. J Exp Med. 1994;180:35–42. doi: 10.1084/jem.180.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Labrecque S, Naor N, Thomson D, Matlashewski G. Analysis of the anti-p53 antibody response in cancer patients. Cancer Res. 1993;53:3468–3471. [PubMed] [Google Scholar]
- 11.Disis ML, Calenoff E, McLaughlin G, Murphy AE, Chen W, Groner B, Jeschke M, Lydon N, McGlynn E, Livingston RB, et al. Existent T-cell and antibody immunity to HER-2/neu protein in patients with breast cancer. Cancer Res. 1994;54:16–20. [PubMed] [Google Scholar]
- 12.Boon T, Coulie PG, Van den Eynde B. Tumor antigens recognized by T cells. Immunol Today. 1997;18:267–268. doi: 10.1016/S0167-5699(97)80020-5. [DOI] [PubMed] [Google Scholar]
- 13.Chen YT, Güre AO, Tsang S, Stockert E, Jäger E, Knuth A, Old LJ. Identification of multiple cancer/testis antigens by allogeneic antibody screening of a melanoma cell line library. Proc Natl Acad Sci USA. 1998;95:6919–6923. doi: 10.1073/pnas.95.12.6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van der Bruggen P, Traversari C, Chomez P, Lurquin C, De Plaen E, Van den Eynde BJ, Knuth A, Boon T. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. J Immunol. 2007;178:2617–2621. [PubMed] [Google Scholar]
- 15.Caballero OL, Chen YT. Cancer/testis (CT) antigens: Potential targets for immunotherapy. Cancer Sci. 2009;100:2014–2021. doi: 10.1111/j.1349-7006.2009.01303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sahin U, Türeci O, Schmitt H, Cochlovius B, Johannes T, Schmits R, Stenner F, Luo G, Schobert I, Pfreundschuh M. Human neoplasms elicit multiple specific immune responses in the autologous host. Proc Natl Acad Sci USA. 1995;92:11810–11813. doi: 10.1073/pnas.92.25.11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Türeci O, Sahin U, Zwick C, Koslowski M, Seitz G, Pfreundschuh M. Identification of a meiosis-specific protein as a member of the class of cancer/testis antigens. Proc Natl Acad Sci USA. 1998;95:5211–5216. doi: 10.1073/pnas.95.9.5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen YT, Scanlan MJ, Sahin U, Türeci O, Gure AO, Tsang S, Williamson B, Stockert E, Pfreundschuh M, Old LJ. A testicular antigen aberrantly expressed in human cancers detected by autologous antibody screening. Proc Natl Acad Sci USA. 1997;94:1914–1918. doi: 10.1073/pnas.94.5.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee SY, Obata Y, Yoshida M, Stockert E, Williamson B, Jungbluth AA, Chen YT, Old LJ, Scanlan MJ. Immunomic analysis of human sarcoma. Proc Natl Acad Sci USA. 2003;100:2651–2656. doi: 10.1073/pnas.0437972100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song MH, Ha JM, Shin DH, Lee CH, Old L, Lee SY. KP-CoT-23 (CCDC83) is a novel immunogenic cancer/testis antigen in colon cancer. Int J Oncol. 2012;41:1820–1826. doi: 10.3892/ijo.2012.1601. [DOI] [PubMed] [Google Scholar]
- 21.Suri A, Jagadish N, Saini S, Gupta N. Targeting cancer testis antigens for biomarkers and immunotherapy in colorectal cancer: Current status and challenges. World J Gastrointest Oncol. 2015;7:492–502. doi: 10.4251/wjgo.v7.i12.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grah JJ, Katalinic D, Juretic A, Santek F, Samarzija M. Clinical significance of immunohistochemical expression of cancer/testis tumor-associated antigens (MAGE-A1, MAGE-A3/4, NY-ESO-1) in patients with non-small cell lung cancer. Tumori. 2014;100:60–68. doi: 10.1700/1430.15817. [DOI] [PubMed] [Google Scholar]
- 23.Sigalotti L, Coral S, Altomonte M, Natali L, Gaudino G, Cacciotti P, Libener R, Colizzi F, Vianale G, Martini F, et al. Cancer testis antigens expression in mesothelioma: Role of DNA methylation and bioimmunotherapeutic implications. Br J Cancer. 2002;86:979–982. doi: 10.1038/sj.bjc.6600174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaykalova DA, Vatapalli R, Wei Y, Tsai HL, Wang H, Zhang C, Hennessey PT, Guo T, Tan M, Li R, et al. Outlier analysis defines zinc finger gene family DNA methylation in tumors and saliva of head and neck cancer patients. PLoS One. 2015;10:e0142148. doi: 10.1371/journal.pone.0142148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hentati A, Bejaoui K, Pericak-Vance MA, Hentati F, Speer MC, Hung WY, Figlewicz DA, Haines J, Rimmler J, Ben Hamida C, et al. Linkage of recessive familial amyotrophic lateral sclerosis to chromosome 2q33-q35. Nat Genet. 1994;7:425–428. doi: 10.1038/ng0794-425. [DOI] [PubMed] [Google Scholar]
- 26.Kiyokawa H, Ray D. In vivo roles of CDC25 phosphatases: Biological insight into the anti-cancer therapeutic targets. Anticancer Agents Med Chem. 2008;8:832–836. doi: 10.2174/187152008786847693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galaktionov K, Lee AK, Eckstein J, Draetta G, Meckler J, Loda M, Beach D. CDC25 phosphatases as potential human oncogenes. Science. 1995;269:1575–1577. doi: 10.1126/science.7667636. [DOI] [PubMed] [Google Scholar]
- 28.Kristjánsdóttir K, Rudolph J. Cdc25 phosphatases and cancer. Chem Biol. 2004;11:1043–1051. doi: 10.1016/j.chembiol.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 29.Kim J, Singh AK, Takata Y, Lin K, Shen J, Lu Y, Kerenyi MA, Orkin SH, Chen T. LSD1 is essential for oocyte meiotic progression by regulating CDC25B expression in mice. Nat Commun. 2015;6:10116. doi: 10.1038/ncomms10116. [DOI] [PMC free article] [PubMed] [Google Scholar]