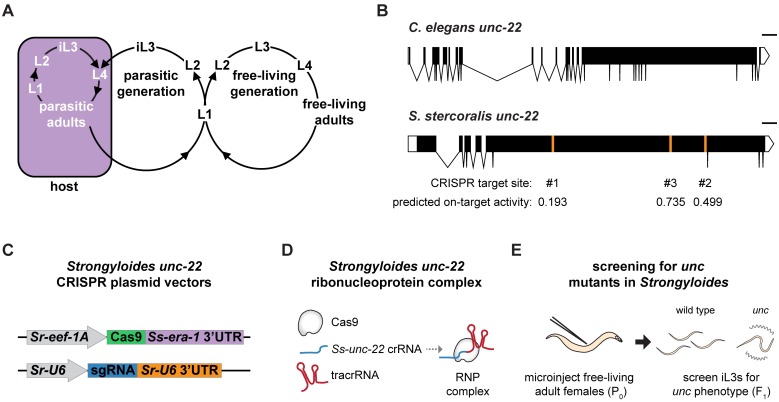
Fig 1. A strategy for targeted mutagenesis in S. stercoralis using CRISPR-Cas9.
(A) The life cycle of S. stercoralis. iL3s enter hosts by skin penetration. The nematodes then develop into parasitic adults, which reside and reproduce in the small intestine. Their progeny exit the host in feces and develop into either iL3s or free-living adults. The free-living adults mate and reproduce in the environment, and all of their progeny develop into iL3s. Thus, S. stercoralis can develop through a single generation outside the host [10]. S. stercoralis can also complete its life cycle within a single host [9]. L1-L4 = 1st-4th larval stages. Adapted from Gang and Hallem, 2016 [10]. (B) The unc-22 genes of C. elegans and S. stercoralis. The Ss-unc-22 gene structure depicted is based on the gene prediction from WormBase ParaSite [24,47]. The CRISPR target sites tested and their predicted on-target activity scores are indicated [50]. Scale bar = 1 kb. (C) Plasmid vectors for the expression of Cas9 and sgRNA in S. stercoralis. (D) RNP complex assembly. Cas9 protein, crRNA targeting Ss-unc-22, and tracrRNA are incubated in vitro to form RNP complexes [20]. (E) Strategy for targeted mutagenesis in S. stercoralis. Plasmid vectors or RNP complexes were introduced into developing eggs by gonadal microinjection of free-living adult females. F1 iL3 progeny were screened for unc phenotypes, putatively resulting from mutation of Ss-unc-22.