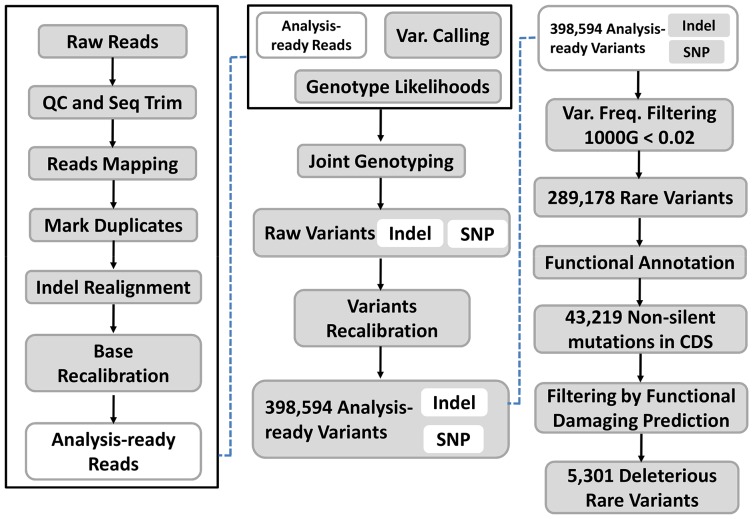
Figure 2. Analysis of whole-exome sequencing data.
We identified 398,594 SNVs. There were 289,178 rare SNVs in 2,034 genes (MAF<0.02, in the 1000 genomes project (http://1000genomes.org)) and 43,219 non-silent mutations in CDS regions. Functional analysis indicated there were 5,301 rare variants predicted to be deleterious. MAF for each SNV was calculated according to the guidline from https://www.ncbi.nlm.nih.gov/projects/SNP/docs/rs_attributes.html.