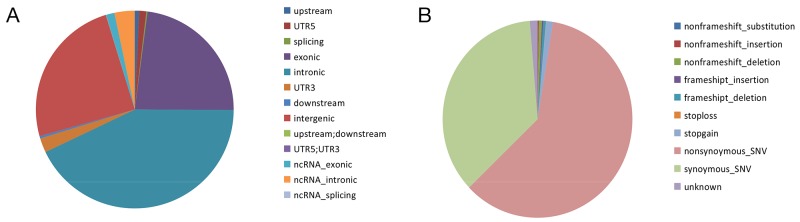
Figure 3. Distribution and functional analysisof rare SNVs.
(A) Of the 289,178 rare SNVs identified, there were 2,035 SNVs in upstream regions; 3,367 SNVs in 5’UTRs; 560 SNVs in splice sites; 66,536 SNVs in exons; 123,938 SNVs in introns; 6,776 SNVs in 3’UTR; 1,085 SNVs in downstream regions; 71,210 SNVs in intergenic regions; 117 SNVs in upstream/downstream regions; 6 SNVs in 5’UTR/3’UTRs; 3,923 SNVs in ncRNA exons; 9,591 SNVs in ncRNA introns; 34 SNVs in ncRNA splice sites. (B) Functional analysis of the 43,219 SNVs inCDS regions revealed 7 SNVs that resulted in non-frameshift substitutions; 127 SNVs that resulted in non-frameshift insertions; 356 SNVs that resulted in non-frameshift deletions; 142 SNVs that resulted in frameshift insertions; 306 SNVs that resulted in frameshift deletions; 26 stop-loss SNVs; 764 stop-gain SNVs; 40,064 non-synonymous SNVs; 23,877 synonymous SNVs; and 867 SNVs of unknown consequence.