SUMMARY
Polymerization and phase separation of proteins containing low-complexity (LC) domains are important factors in gene expression, mRNA processing and trafficking, and localization of translation. We have used solid state nuclear magnetic resonance methods to characterize the molecular structure of self-assembling fibrils formed by the LC domain of the fused in sarcoma (FUS) RNA-binding protein. From the 214-residue LC domain of FUS (FUS-LC), a segment of only 57 residues forms the fibril core, while other segments remain dynamically disordered. Unlike pathogenic amyloid fibrils, FUS-LC fibrils lack hydrophobic interactions within the core and are not polymorphic at the molecular structural level. Phosphorylation of core-forming residues by DNA-dependent protein kinase blocks binding of soluble FUS-LC to FUS-LC hydrogels and dissolves phase-separated, liquid-like FUS-LC droplets. These studies offer a structural basis for understanding LC domain self-assembly, phase separation, and regulation by post-translational modification.
Keywords: FUS, low-complexity sequence, labile cross-β polymer, solid state nuclear magnetic resonance, amyloid structure, liquid droplet, liquid-liquid phase separation, electron microscopy, amyotrophic lateral sclerosis, neurodegeneration
INTRODUCTION
Proteins containing low-complexity (LC) domains perform a variety of functions in essential cellular processes, including gene expression, mRNA processing, nuclear export and import, and localized translation (Dyson and Wright, 2005; Kiebler and Bassell, 2006; Lamond and Spector, 2003; Liu et al., 2006; Strambio-De-Castillia et al., 2010). LC domains can be hundreds of residues in length, have simple amino acid compositions, and have often been considered to function as intrinsically disordered monomers. Recently, several studies have given evidence that self-assembly of LC domains may underlie the formation of membrane-less intracellular puncta (Banani et al., 2017). Additionally, aberrant self-assembly of LC domains has been implicated in a variety of neurodegenerative diseases (Alberti and Hyman, 2016; Harrison and Shorter, 2017; Taylor et al., 2016).
Incubation of purified LC domains in vitro can lead to the formation of phase-separated, liquid-like droplets capable of maturing into a hydrogel state (Elbaum-Garfinkle et al., 2015; Kato et al., 2012; Lin et al., 2016; Murakami et al., 2015; Nott et al., 2015; Patel et al., 2015). LC polypeptides in hydrogels are non-covalently polymerized into morphologically homogeneous, amyloid-like fibrils. Data from both x-ray fiber diffraction and electron microscopy suggest that LC domain fibrils contain cross-β structures similar to those in pathogenic amyloid and prion fibrils (Kato et al., 2012). However, unlike pathogenic fibrils, LC domain fibrils are readily disassembled by dilution, temperature changes, or mild treatment with detergent (Kato et al., 2012; Lin et al., 2016; Murakami et al., 2015; Shi et al., 2017)
The state of interaction of LC polypeptides within liquid-like droplets has been controversial. Several investigators have reported that LC polypeptides remain in a fully disordered conformation within these droplets (Burke et al., 2015; Feric et al., 2016; Nott et al., 2015; Patel et al., 2015; Saha et al., 2016). In contrast, other studies provide evidence that LC polypeptides adopt similar conformations in liquid-like droplets, hydrogel polymers, and mammalian cell nuclei (Xiang et al., 2015).
The structural basis for polymerization of LC domains is largely unexplored. Although detailed molecular structural models for disease-associated amyloid fibrils, especially fibrils formed by amyloid-β (Aβ) peptides (Bertini et al., 2011; Colvin et al., 2016; Lu et al., 2013; Paravastu et al., 2008; Schutz et al., 2015; Walti et al., 2016; Xiao et al., 2015) and α-synuclein (Tuttle et al., 2016), have been developed from solid state nuclear magnetic resonance (NMR) spectroscopy, the extent to which LC domain polymers resemble pathogenic amyloid fibrils at the molecular level has been unclear. In particular, LC sequences differ substantially in amino acid composition, length, and sequence repetition from polypeptides such as Aβ and α-synuclein.
Here we report results from solid state NMR experiments to characterize the molecular structure of fibrils formed by the LC domain of the fused in sarcoma (FUS) RNA-binding protein (FUS-LC). Despite the repetitive nature of the FUS-LC sequence, we find that the structured core of FUS-LC fibrils is formed reproducibly by a specific 57-residue segment in the N-terminal half. Other segments, especially in the C-terminal half, remain dynamically disordered. Interestingly, the amino acid composition of the core-forming segment of FUS-LC is not much different from the overall composition, showing that subtle variations in sequence can distinguish structure-forming segments from dynamically disordered segments within an LC polymer. From a large set of experimental restraints, we develop a detailed structural model for the FUS-LC fibril core, which may serve as a basis for understanding both biophysical and biologic properties of LC domains in general.
We also report results from experiments that examine the effects of phosphorylation of Ser and Thr sidechains by DNA-dependent protein kinase (DNA-PK) on FUS-LC self-assembly (binding to preformed FUS-LC hydrogels) and phase separation of FUS-LC droplets. The DNA-PK experiments reveal site-specific variations in the effects of phosphorylation that correlate with the fibril core-forming segment identified by solid state NMR experiments. The site-specificity of effects on droplet formation argues in favor of a molecular structural contribution to LC domain phase separation, as well as a similarity between FUS-LC conformations in fibrils and droplets.
RESULTS
Fibril formation by FUS-LC
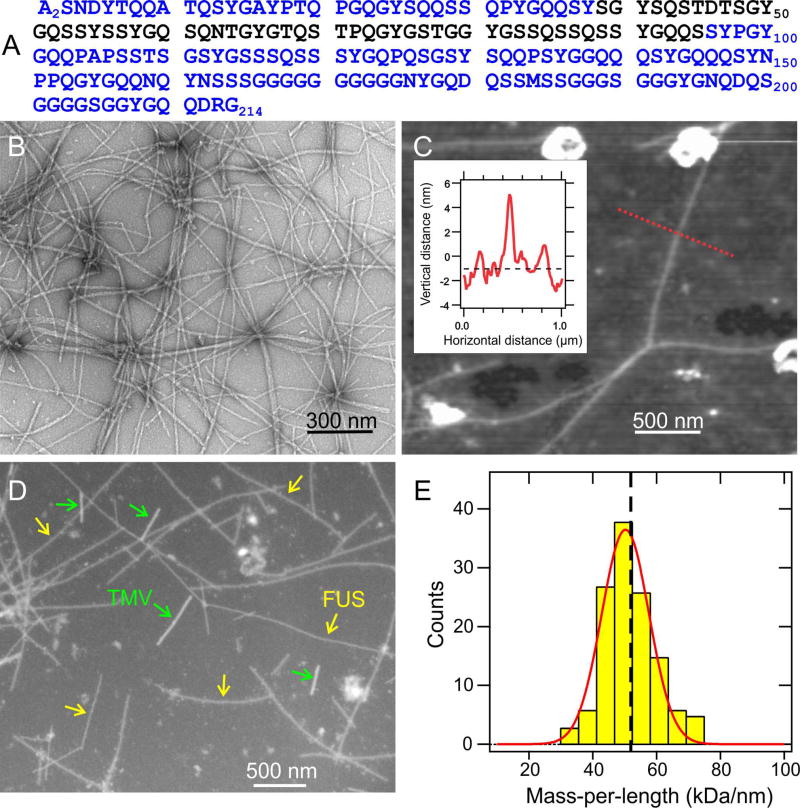
His-tagged FUS-LC, bearing the amino acid sequence in Fig. 1A, was expressed in E. coli and purified by Ni2+ affinity chromatography under denaturing conditions. Polymerization was initiated by concentrating the protein to approximately 30 µM, removing denaturant by dialysis, and incubating at room temperature. Fibrils were also prepared by seeded growth (see Method Details). As shown in Fig. 1B, negatively-stained FUS-LC fibrils have the straight, unbranched appearance of typical amyloid fibrils in transmission electron microscope (TEM) images. Atomic force microscope (AFM) images indicate uniform fibril diameters of 5.5 ± 0.7 nm (Fig. 1C). FUS-LC fibrils also exhibit typical amyloid dye-binding properties (Fig. S1A,B).
Fig. 1. Fibril formation by the low-complexity domain of FUS.
(A) Human FUS-LC sequence. Experiments were performed on samples with an additional N-terminal His tag, bearing the sequence MSYYHHHHHHDYDIPTTENLYFQGAMDP. (B) TEM image of negatively-stained FUS-LC fibrils. (C) AFM image of FUS-LC fibrils adsorbed to mica, with fibril heights indicating diameters of 5.5 ± 0.7 nm. Inset shows the height profile along the dotted red line. (D) Dark-field TEM image of unstained FUS-LC fibrils. Tobacco mosaic virus (TMV) particles are included as mass-per-length (MPL) standards for image intensity calibration. (E) MPL histogram obtained from multiple dark-field images. Red line is a Gaussian fit, centered at 50 kDa/nm, with 11 kDa/nm full-width-at-half-maximum. Vertical dashed line indicates the MPL value expected for a single cross-β structural unit with 0.48 nm intermolecular spacing. See also Figs. S1 and S7.
Measurements of the mass-per-length (MPL) of individual fibrils by dark-field TEM (Chen et al., 2009) (Fig. 1D and Fig. S1C) yielded the MPL histogram in Fig. 1E. The histogram was fit with a single Gaussian distribution centered at MPL = 50 ± 2 kDa/nm, in good agreement with the 51.9–53.0 kDa/nm value expected from a single cross-β structural unit comprised of monomers with the 24.9 kDa molecular weight of His-tagged FUS-LC and the 0.47–0.48 nm intermolecular spacing of a standard β-sheet. The width of the MPL histogram (11 kDa/nm full-width-at-half-maximum) arises from random background intensity variations in the dark-field images (see Fig. S1D).
NMR spectra indicate a structurally ordered core in FUS-LC fibrils, coexisting with dynamically disordered segments
NMR measurements were performed initially on uniformly 15N,13C-labeled FUS-LC (U-FUS-LC) fibrils, pelleted by ultracentrifugation and loaded directly into magic-angle spinning (MAS) rotors without lyophilization or drying. Measurements under conditions appropriate for solid state NMR, especially the use of cross-polarization (CP) driven by magnetic dipole-dipole couplings for nuclear spin polarization transfers (Pines et al., 1973) and high-power 1H decoupling (Bennett et al., 1995), select NMR signals from residues that are immobilized in the fibrils. Conversely, measurements under conditions similar to those in solution NMR spectroscopy, especially the use of the INEPT (Insensitive Nuclei Enhanced by Polarization Transfer) technique for polarization transfers driven by scalar couplings (Morris and Freeman, 1979) and low-power 1H decoupling, select NMR signals from residues that undergo rapid and nearly isotropic motion.
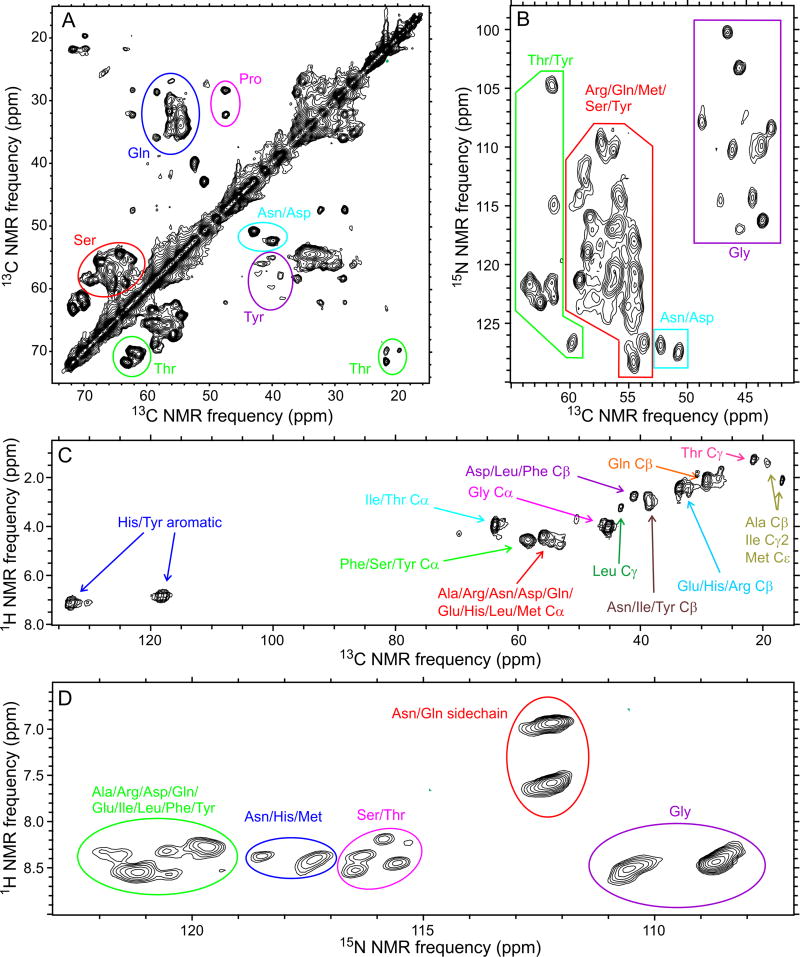
Two-dimensional (2D) 13C-13C and 15N-13C spectra of U-FUS-LC fibrils, obtained with solid state NMR (i.e., CP-based) conditions, contain strong, sharp crosspeaks attributable to approximately 50 residues, including ten Gly, ten Ser, five Thr, one Pro, and two Asp/Asn residues (Fig. 2A,B). No Ala signals were detected. Crosspeak patterns were reproducible over multiple independent sample preparations, with minor variations in intensities of weaker crosspeaks, indicating a single predominant FUS-LC fibril structure without the variable polymorphism that has been observed in solid state NMR studies of other amyloid fibrils, even under a single set of growth conditions (Heise et al., 2005; Paravastu et al., 2008; Xiao et al., 2015).
Fig. 2. Coexistence of structural order and dynamic disorder within FUS-LC fibrils.
(A,B) 2D 13C-13C and 15N-13C NMR spectra of U-FUS-LC fibrils, recorded under conditions that select for signals from immobilized, structurally ordered segments. (C,D) 2D 1H- 13C and 1H-15N NMR spectra of U-FUS-LC fibrils, recorded under conditions that select for signals from highly flexible, dynamically disordered segments. Residue-type assignments are based on characteristic chemical shift ranges. Contour levels increase by successive factors of 1.40, 1.35, 1.80, and 1.40 in (A), (B), (C), and (D), respectively. See also Figs. S2 and S3 and Table S3.
Four types of three-dimensional (3D) solid state NMR spectra of U-FUS-LC fibrils were also recorded (Fig. S2). Crosspeak signals attributable to 49–57 residues were detected in each of the 3D spectra with adequate signal-to-noise for subsequent analysis (55 in NCACX, 49 in NCOCX, 57 in CONCA, 49 in CANCX spectra). Differences in the number of detectable residues in different 3D spectra arise from differences in polarization transfer pathways, spectral resolution, and overall signal-to-noise. From the 2D and 3D solid state NMR spectra, it is clear that the core of FUS-LC fibrils contains fewer than 60 structurally ordered residues, although the full sequence (without the His tag) contains 214 residues.
NMR spectra of U-FUS-LC fibrils were also obtained under solution NMR conditions (Fig. 2C,D). 2D 1H-13C and 1H-15N INEPT spectra show strong crosspeaks from nearly all amino acid types in FUS-LC, with 1H and 13C chemical shifts close to random coil values. The average transverse 15N spin relaxation time (T2 value) for these signals was 73 ± 2 ms, as determined by a spin echo decay after 1H-15N INEPT. Thus, certain segments remain highly flexible in FUS-LC fibrils, executing nearly isotropic motions on sub-microsecond time scales.
Identification of the core-forming segments
Assignment of signals in 3D solid state NMR spectra of U-FUS-LC fibrils to specific amino acid sites allows us to identify the immobilized, core-forming segments. However, the repetitive nature of the FUS-LC sequence and the fact that less than 25% of the sequence contributes to these spectra makes it difficult to establish the uniqueness of assignments obtained by conventional manual methods. Therefore, we used a computationally-aided approach based on the MCASSIGN algorithm (Hu et al., 2011), which has been applied previously in solid state NMR studies of amyloid fibrils and viral capsid assemblies (Bayro et al., 2014; Lu et al., 2013). Briefly, the MCASSIGN algorithm uses a Monte Carlo/simulated annealing approach to assign sets of crosspeak signals from each 3D spectrum in a manner that is consistent with the amino acid sequence, with manual residue-type assignments for each signal, and with the required connections (i.e., chemical shift matches) among the 3D spectra. Results from multiple MCASSIGN runs can be compared, allowing signals with assignments that are uniquely defined by the data to be distinguished from signals with multiple possible assignments.
Two independent sets of MCASSIGN calculations, using somewhat different approaches and input files, resulted in unique signal assignments to residues 39–40, 44–54, and 63–95 (Fig. S3A; Table S1; Method Details). We therefore conclude that residues 39–95 constitute the structurally ordered core of FUS-LC fibrils, with partial disorder or enhanced dynamics in residues 55–62 that precludes detection or unambiguous assignment of solid state NMR signals to these sites.
Crosspeak amplitudes for different residues varied significantly. For example, in the 3D CONCA spectrum of U-FUS-LC fibrils, assigned crosspeaks from 43 residues have signal-to-noise ratios above 5.0, but the amplitudes of these crosspeaks vary by a factor of 5.2 (Fig. S3B). The observed variations in crosspeak amplitudes suggest a distribution of motional amplitudes or time scales, even within the fibril core.
Certain signals in the 3D spectra could not be assigned uniquely (Table S2, Fig. S3C). These signals have relatively low signal-to-noise in most cases, and apparently arise from individual residues or short segments that are sufficiently immobile to contribute to some 3D spectra but are flanked by more mobile residues, thereby preventing sequential signal assignment. Thus, certain segments outside of residues 39–95 are partially ordered, perhaps by loose association with the FUS-LC fibril core or by inter-fibril interactions.
Confirmation by segmental isotopic labeling
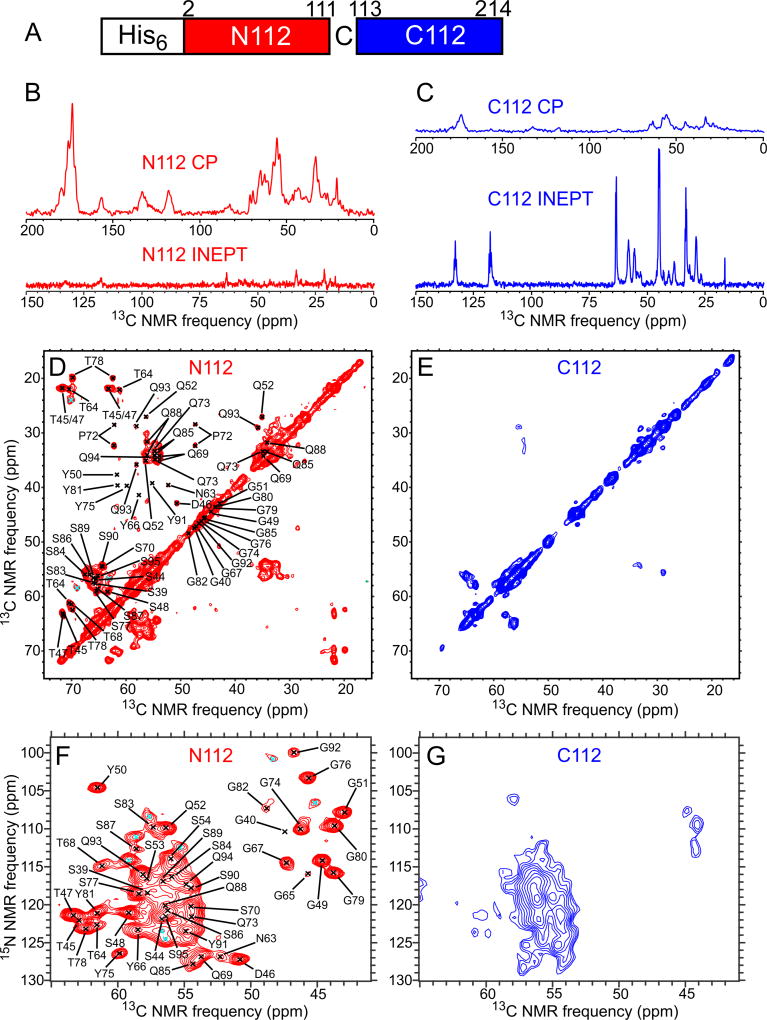
As an additional test of the identity of FUS-LC fibril core, samples were prepared with uniform 15N,13C-labeling of either N-terminal or C-terminal segments of the FUS-LC sequence, using native chemical ligation of labeled and unlabeled segments (Method Details; Fig. S4). We introduced a single Cys substitution at the junction between labeled and unlabeled segments (Fig. 3A) as required for the ligation reaction (Shah et al., 2012).
Fig. 3. Segmental isotopic labeling indicates a fibril core-forming segment in the N-terminal half of FUS-LC.
(A) Schematic representation of the 15N,13C-labeled segments in N112-FUS-LC and C112-FUS-LC, with a Ser-to-Cys substitution at residue 112. (B) 1D 13C NMR spectra of N112-FUS-LC fibrils. Spectra obtained with either 1H-13C CP or 1H-13C INEPT are plotted with the same vertical scale, after correcting for differences in signal averaging. (C) Same as panel B, but for CI 12-FUS-LC fibrils. Spectra obtained with CP show signals from immobilized, structurally ordered sites, while spectra obtained with INEPT show signals from highly flexible, dynamically disordered sites. (D,E) 2D 13C-13C NMR spectra of N112- and CI 12-FUS-LC fibrils, recorded with 1H-13C CP. (F,G) 2D 15N-13C NMR spectra of N112- and C112-FUS-LC fibrils, recorded with 1H-15N CP. Contour levels increase by successive factors of 1.20 in panels D–G. Black crosses in panels D and F indicate crosspeak positions from residues with definite chemical shift assignments. Signals from some of these residues are too weak to be detected in these 2D spectra. Cyan crosses indicate crosspeaks from signals that could not be assigned definitely to specific residues. See also Figs. S4 and S5 and Tables S1 and S2.
Figs. 3B and 3C show 1D 13C NMR spectra of fibrils with a Ser112-Cys substitution and with labeling of either residues 1–111 (N112-FUS-LC) or residues 112–214 (C112-FUS-LC). For N112-FUS-LC fibrils, the ratio of the total 13C NMR signal amplitude from aliphatic sites observed under solid state NMR conditions to that observed under solution NMR conditions is 13 ± 1. For C112-FUS-LC fibrils, this ratio is 0.47 ± 0.02. Thus, most immobilized sites are in residues 1–111. Figs. 3D–G show 2D solid state NMR spectra of N112-FUS-LC and C112-FUS-LC fibrils. 2D spectra of N112-FUS-LC fibrils are nearly identical to corresponding spectra of U-FUS-LC fibrils (Fig. 2A,B), confirming that the core-forming segment resides in residues 1–111. 3D CONCA and NCACX spectra of N112-FUS-LC fibrils were also recorded and used to confirm signal assignments (Method Details).
2D solid state NMR spectra of C112-FUS-LC fibrils show relatively weak and broad signals, with few of the sharp crosspeaks that are observed in the corresponding spectra of U-FUS-LC fibrils. Solid state NMR signals from C112-FUS-LC fibrils are attributable to partially ordered residues outside the fibril core, as discussed above. Fibrils were also prepared with a Gln60-Cys substitution and with labeling of either residues 1–59 (N60-FUS-LC) or residues 60–214 (C60-FUS-LC) (Fig. S5A). For N60-FUS-LC fibrils, the ratio of the total 13C NMR signal amplitude from aliphatic sites observed under solid state NMR conditions to that observed under solution NMR conditions is 8.2 ± 0.8. For C60-FUS-LC fibrils, this ratio is 1.26 ± 0.06 (Fig. S5B,C). 2D solid state NMR spectra of these samples are also consistent with signal assignments from 3D spectra of U-FUS-LC fibrils (Fig. S5D–G).
Structural model for the FUS-LC fibril core
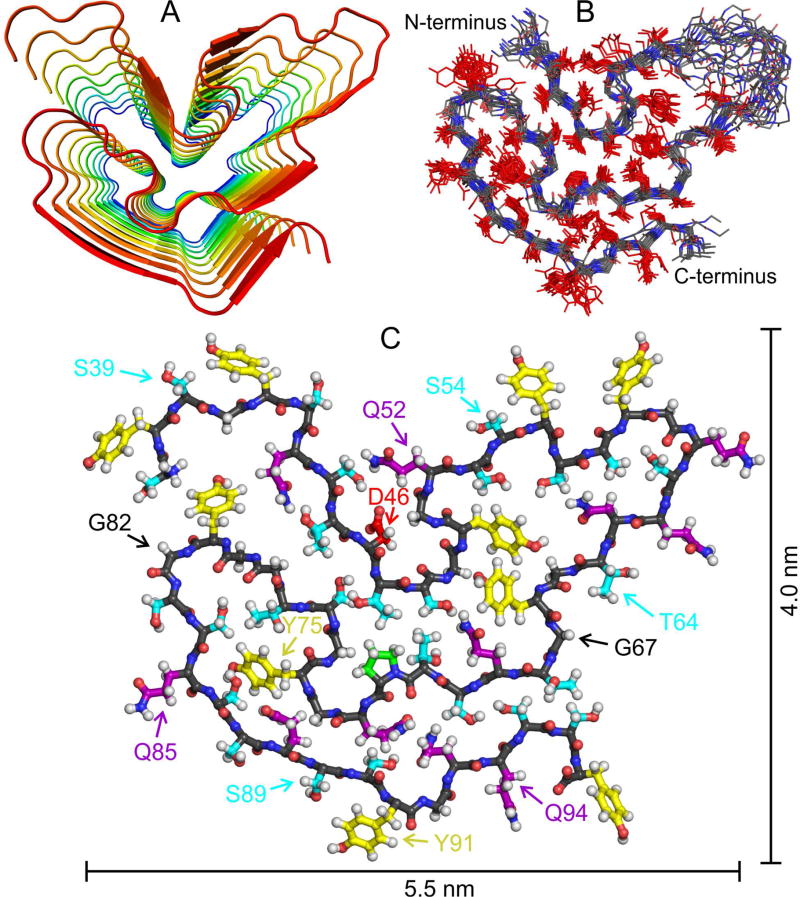
Development of a detailed molecular structural model for the FUS-LC fibril core requires experimental restraints on intermolecular alignment, backbone conformation, and inter-residue contacts. Structures were calculated with the Xplor-NIH program (Schwieters et al., 2006), using the following restraints: (i) MPL data in Fig. 1 and the observation of a single set of solid state NMR chemical shifts, implying that FUS-LC fibrils contain a single cross-β unit in which all molecules have equivalent conformations and structural environments; (ii) Predictions of backbone φ and ψ torsion angles from 15N and 13C chemical shifts by the TALOS-N program (Shen and Bax, 2013) (Table S1, Fig. S6A); (iii) Measurements of intermolecular 13C-13C magnetic dipole-dipole couplings with the PITHIRDS-CT technique (Tycko, 2007) on FUS-LC fibrils that were 13C-labeled at backbone carbonyl sites of either all Tyr residues or all Thr residues. These data indicate intermolecular 13C-13C distances of approximately 0.47 ± 0.02 nm (Fig. S6B), implying an in-register parallel intermolecular alignment within the cross-β core; (iv) Measurements of 15N-15N dipole-dipole couplings among nearest-neighbor backbone nitrogen sites with the 15N-BARE technique (Hu et al., 2012) on U-FUS-LC fibrils and on FUS-LC fibrils that were uniformly 15N-labeled and partially 13C-labeled (Castellani et al., 2003; Hong and Jakes, 1999), using 2-13C-glycerol as the carbon source (2-Glyc-FUS-LC; Fig. S6C); (v) Measurements of long-range inter-residue crosspeaks in 2D 13C-13C and 3D NCACX and NCOCX spectra of 2-Glyc-FUS-LC fibrils and FUS-LC fibrils that were uniformly 15N-labeled and partially 13C-labeled, using 1,3-13C-glycerol as the carbon source (1,3-Glyc-FUS-LC; Fig. S6D,E). Xplor-NIH calculations were performed on nine copies of residues 37–97, resulting in final models that represent a fibril segment with a length of approximately 3.8 nm (Method Details). Restraints and structure statistics are summarized in Tables S4 and S5. The final bundle of 20 structural models was deposited in the Protein Data Bank with code 5W3N.
As shown in Fig. 4, the overall backbone conformation of residues 43–95 and the orientations of sidechains for residues 44–52 and 65–95 are well determined by the experimental restraints. (Sidechain orientations are reversed for residues 86–90 and 93–95 in one out of 20 models.) Sidechain orientations and backbone torsion angles are not determined for residues 53–64, reflecting the absence of chemical shift assignments for residues 55–62. These residues are likely to be partially dynamic in FUS-LC fibrils. Sidechain torsion angles are not restrained by the existing data, so variations in sidechain conformations do not necessarily indicate real static or dynamic disorder.
Fig. 4. Structural model for the FUS-LC fibril core.
(A) Cartoon representation of residues 37–97, viewed down the fibril growth axis, illustrating the overall fold, the in-register parallel alignment of FUS-LC monomers, and the absence of β-strand segments longer than six residues. (B) Superposition of the central copy of residues 37–97 in twenty low-energy models. The backbone conformation and sidechain orientations (but not sidechain conformations) are well determined by experimental restraints. (C) Monomer conformation of the lowest-energy model, with backbone carbons in black. Sidechain carbons are colored by residue type, with Ser and Thr in cyan, Tyr in yellow, Gln and Asn in purple, Pro in green, and Asp in red. See also Fig. S6, Tables S4 and S5, and Movies S1 and S2.
Segments that form β-strands of a cross-β motif in all 20 models, defined by the characteristic alignment of backbone carbonyl groups with the fibril growth axis, include residues 44–46, 52–54, 62–64, 67–70, 85–90, and 93–95. It appears that much of the structurally ordered core of FUS-LC fibrils (roughly 50% of residues with assigned solid state NMR signals) does not participate in the intermolecular hydrogen bonding interactions that help stabilize a cross-β structure.
Phosphorylation-mediated effects on hydrogel binding and liquid-like droplet formation by FUS-LC
The LC domain of FUS can be phosphorylated by DNA-dependent protein kinase (DNA-PK) upon DNA damage (Gardiner et al., 2008). If pre-exposed to DNA-PK, a fusion protein linking FUS-LC to green fluorescent protein (GFP) is blocked from binding to hydrogels composed of FUS-LC linked to mCherry (Han et al., 2012). Although, liquid-like droplet formation by the LC domain of FUS has been studied (Burke et al., 2015; Lin et al., 2015; Murakami et al., 2015; Patel et al., 2015), effects of phosphorylation on droplet formation have not been described previously.
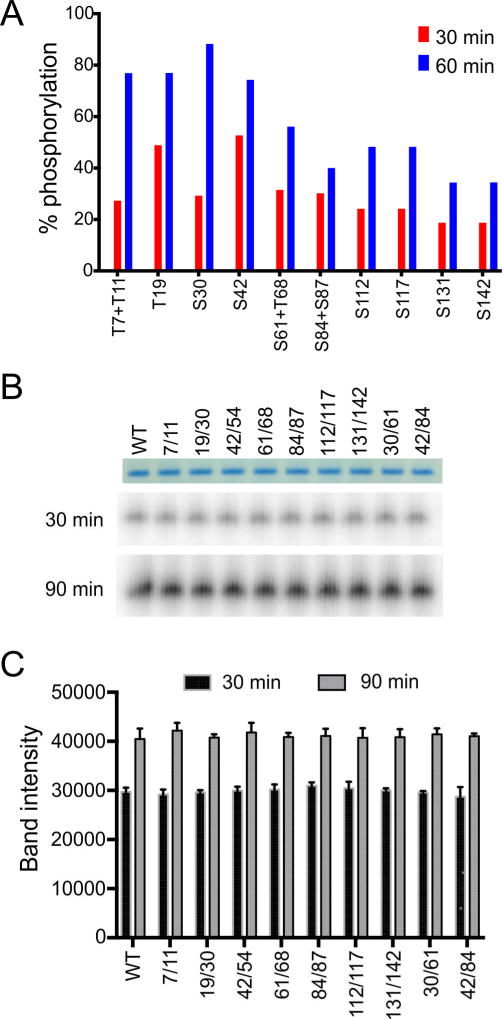
In living cells, phosphorylation of the LC domain of FUS has been reported to correlate with translocation of the protein from the cell nucleus to the cytoplasm upon DNA damage, coupled with a distinct alteration in its pattern of migration as deduced by SDS-PAGE (Deng et al., 2014). It has also been reported that all phosphorylation of the FUS protein incurred upon DNA damage is localized to the LC domain (Deng et al., 2014). To establish a comprehensive map of DNA-PK phosphorylation sites, a purified fusion protein linking FUS-LC to GFP was incubated with DNA-PK in the presence of ATP. The fusion protein was then digested with chymotrypsin and subjected to proteomic analysis to map phosphorylation sites via mass spectrometry (see Method Details). These efforts led to the identification of 14 phosphorylation sites, including Ser residues 30, 42, 54, 61, 84, 87, 112, 117, 131 and 142, and Thr residues 7, 11, 19, and 68 (Fig. 5A). The level of phosphorylation of individual sites increased proportionally upon incubation with DNA-PK and ATP from 30 to 60 min, and the ultimate level of phosphorylation for the individual sites, or closely spaced pairs of sites, ranged from roughly 30% to 80%.
Fig. 5. Phosphorylation of wild-type and double-mutant FUS-LC by DNA-PK.
(A) Quantification by mass spectrometry of phosphorylation at specific Ser and Thr sites in wild-type GFP:FUS-LC, after treatment with DNA-PK and ATP for indicated time periods. (B,C) Analysis of total phosphorylation levels of wild-type FUS-LC and double phosphorylation-site mutants of FUS-LC analyzed by autoradiography. No significant differences in phosphorylation levels were detected.
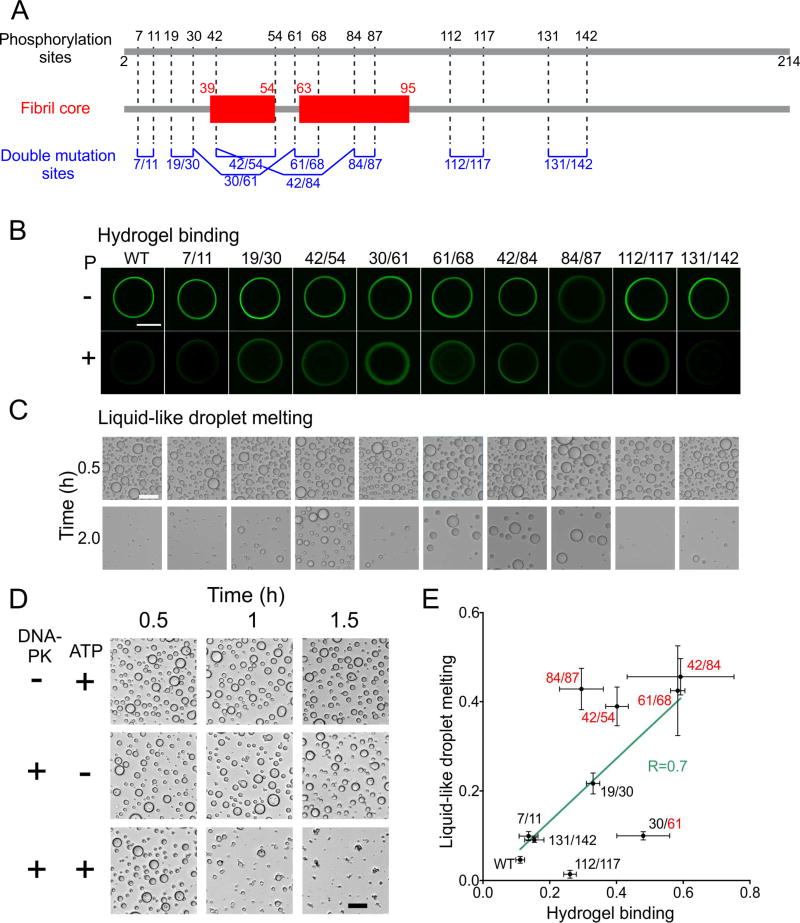
As a means of blocking phosphorylation at specific sites, pairs of Ser and Thr residues were simultaneously mutated to Ala, generating nine double mutants (Fig. 6A). All double mutants were phosphorylated to the same degree as wild-type FUS-LC (Fig. 5B,C), showing that none of the mutated sites dominates the ultimate level of DNA-PK-mediated phosphorylation. All double mutants were compared in hydrogel binding assays, using fusion proteins in which double mutant FUS-LC was linked to GFP (Fig. 6B). In the absence of phosphorylation, all but one of the double mutants were observed to bind to mCherry:FUS hydrogel droplets to an equivalent degree as the wild-type FUS-LC:GFP fusion protein. Hydrogel binding by the S84A/S87A mutant was observed to be reduced relative to wild-type protein by approximately 50%. According to the structural model for FUS-LC fibrils, S84 can form a hydrogen bond network with Y75 and T78 (Fig. 4C). This hydrogen bond network may be important to stabilize the loop between residues 74 and 87. Since both S84 and S87 are located in this loop, disruption of the loop conformation may impair FUS-LC fibril formation.
Fig. 6. Effects of phosphorylation-site mutations on hydrogel binding and liquid-like droplet dissolution.
(A) DNA-PK phosphorylation sites in FUS-LC identified by mass spectrometry (top), structurally ordered segments in FUS-LC fibrils identified by solid state NMR (middle), and Ala substitution sites for FUS-LC double mutants (bottom). (B) Fluorescence microscope images of wild-type and double-mutant GFP:FUS-LC from hydogel binding assays with and without DNA-PK phosphorylation (bottom and top rows, respectively). Methods to quantify hydrogel binding by test proteins have been discussed previously (Kato et al., 2012; Xiang et al., 2015). Abrogation of hydrogel binding by phosphorylation is reduced for certain double mutants. (C) Optical images of liquid-like droplets formed by wild-type and double-mutant FUS-LC in the presence of DNA-PK and ATP, recorded at T = 0.5 h and 2.0 h after initiation of droplet formation. Droplet melting caused by gradual phosphorylation is reduced for certain double mutants. Scale bars in panels B and C are 500 µm and 50 µm, respectively. (D) Visualization of liquid-like droplet formation by wild-type FUS-LC at indicated time points after initial preparation of the protein solution. Droplets dissolved progressively in the presence of both DNA-PK and ATP, but were stable in the absence of either component. Scale bar is 50 µm. (E) Correlation plot for effects of DNA-PK phosphorylation on hydrogel binding and liquid-like droplet melting by FUS-LC double mutants. The horizontal axis shows ratios of GFP fluorescence intensities from phosphorylated GFP:FUS-LC on mCherry-FUS hydrogels to corresponding intensities from control samples. The vertical axis shows ratios of the total areas of liquid-like droplets of His6-K-FUS-LC remaining at the 2.0 h time point to total areas of liquid-like droplets at the 0.5 h time point. Solid line is a least-squares fit to the experimental points. The correlation coefficient is R = 0.70. See also Table S5.
Phosphorylation by DNA-PK reduced binding of wild-type FUS-LC: GFP to mCherry:FUS hydrogels by a factor of ten (Fig. 6B). Similarly large reductions in hydrogel binding were observed for the T7A/T11A and S131A/S142A mutants, consistent with phosphorylation of residues 7, 11, 131, and 142 having little effect on hydrogel binding by wild-type FUS-LC. Reductions by a factor of approximately three were observed for the S112A/S117A, S84A/S87A, and T19A/S30A mutants. Smaller reductions in hydrogel binding upon phosphorylation were observed for the S42A/S54A, S30A/S61A, S61A/T68A, and S42A/S84A mutants.
The same nine double mutants were also examined in assays scoring for the formation of liquid-like droplets. In this assay, His-tagged FUS-LC was assayed as an isolated, purified polypeptide not linked to GFP (Method Details). Without phosphorylation, all nine double mutants formed droplets in a manner indistinguishable from the wild-type protein. Inclusion of both DNA-PK and ATP in the assay allowed initial droplet formation by wild-type FUS-LC, followed by dissolution of the droplets over a two hour period (Fig. 6C). No such "droplet melting" was observed upon elimination of either ATP or DNA-PK from the reaction mixture (Fig. 6D). For four of the double mutants, droplet melting induced by phosphorylation was strongly impeded (factor of 8–10), including the S42A/S54A, S61A/T68A, S42A/S84A and S84A/S87A double mutants. Droplet melting was partially impeded for the T19A/S30A double mutant and only weakly affected for the T7A/T11A, S30A/S61A, S112A/S117A and S131A/S142A double mutants (Fig. 6C).
Effects of the double Ala substitutions on the ability of DNA-PK to either inhibit hydrogel binding by FUS-LC or melt FUS-LC droplets were correlatively similar (Fig. 6E). Elimination of phosphorylation sites within the core-forming region identified by solid state NMR (i.e., the S42A/S54A, S61A/T68A, and S42A/S84A mutants) strongly reduced the effects of DNA-PK treatment on both hydrogel binding and droplet formation. By contrast, elimination of phosphorylation sites outside the fibril core (i.e., the T7A/T11A, T19A/S30A, S112A/S117A and S131A/S142A mutants) generally had less impact on the effects of DNA-PK treatment in both assays.
In order to quantitatively evaluate these data, we separated mutated residues into two groups according to their effects on the ability of DNAPK to either impede hydrogel binding or dissolve liquid-like droplets. None of the mutations in six different non-core residues were observed to impede the ability of DNA-PK to either inhibit hydrogel binding or dissolve liquidlike droplets. Of eight residues mutated to eliminate sites of phosphorylation that could be interpreted to impede the ability of DNA-PK to inhibit hydrogel binding, six mapped to the fibril core. Likewise, of six residues mutated to eliminate DNAPK phosphorylation sites resulting in impediments in phosphorylation-mediated dissolution of liquid-like droplets, five mapped within the fibril core. The partitioning of core phosphorylation sites in the phenotypic effects of DNA-PK-mediated phosphorylation on the two assays were of substantive statistical significance (Table S5).
DISCUSSION
Summary of NMR results
NMR spectra obtained under conditions that select signals from immobilized, structurally-ordered residues show that the core of FUS-LC fibrils contains only a small subset of the full FUS-LC sequence (Fig. 2A,B). NMR spectra obtained under conditions that select signals from dynamically disordered residues show that a large part of the sequence remains highly flexible (Fig. 2C,D). In principle, the fibril core could be comprised of multiple protein segments that are separated by long disordered loops. However, site-specific assignments of the solid state NMR signals (Figs. S2 and S3; Table S1) indicate that the core-forming segments are localized within residues 39–95, with one partially dynamic loop in residues 55–62. The size of the FUS-LC fibril core (roughly 60 amino acids), approximates the sizes of fibril cores assembled from the LC domains of Nup54 and hnRNPA2, as deduced from N-acetylimidazole footprinting (Shi et al., 2017; Xiang et al., 2015).
Identification of the FUS-LC fibril core is confirmed by NMR spectra of segmentally-labeled FUS-LC fibrils (Figs. 3 and S5). Highly dynamic segments are primarily localized in the C-terminal half of FUS-LC. N-terminal segments before residue 39 may have slower dynamics, as suggested by the weakness of INEPT signals in Figs. 3B and S5B. From a variety of experimental restraints (Fig. S6; Table S4), we developed the structural model for the FUS-LC fibril core shown in Fig. 4. The principal structural features are well determined by the experimental data, including intermolecular alignment, backbone conformation, and sidechain orientations. Sidechain conformations are undetermined.
Movie S1 of Supplemental Information depicts the FUS-LC fibril as revealed by NMR measurements, with its relatively rigid, structurally ordered core surrounded by dynamically disordered N- and C-terminal segments. Future experiments may provide additional insight into quantitative aspects of the motional amplitudes and time scales of these segments, as well as their roles in functional interactions.
Movie S2 of Supplemental Information shows results from molecular dynamics simulations on a short piece of the FUS-LC core structure, consisting of five copies of residues 37–97, in explicit water (see Methods Details). Although water molecules penetrate the core within 10 ns, the core structure remains stable for the 150 ns duration of the simulation. This observation suggests that relatively small clusters of FUS-LC molecules can adopt stable or metastable oligomeric configurations that closely resemble the molecular structure within FUS-LC fibrils.
What determines the fibril core?
Our finding that residues 39–95 form the core of FUS-LC fibrils is not predicted by computational methods that assess propensities for amyloid formation by segments within protein sequences (Fig. S7). Aggrescan, FISH, FoldAmyloid, MetAmyl, and PASTA 2.0 predict a low propensity for amyloid formation across the FUS-LC sequence, presumably because these algorithms emphasize the importance of hydrophobic interactions (de Groot et al., 2012; Emily et al., 2013; Garbuzynskiy et al., 2010; Gasior and Kotulska, 2014; Walsh et al., 2014). ZipperDB, Zyggregator, and WALTZ, which include other physicochemical and structural properties of protein segments (Thompson et al., 2006) (Tartaglia and Vendruscolo, 2008) (Maurer-Stroh et al., 2010), predict that certain short segments of FUS-LC may form amyloidlike structures, but with no consensus on the identities of these segments.
Why then do residues 39–95 form the fibril core? The amino acid composition of residues 39–95 (21.1% Gly, 31.6% Ser, 17.5% Gln, 14.0% Tyr, and 10.5% Thr) is not very different from that of residues 1–214 (21.8% Gly, 23.8% Ser, 20.1% Gln, 12.6% Tyr, and 4.7% Thr). The amino acid composition of residues 39–95 is also not very different from that of residues 96–165 (15.7% Gly, 28.6% Ser, 24.2% Gln, 14.3% Tyr, and 1.4% Thr), apart from the absence of Thr residues after T109. Apparently, subtle differences in local sequences are important. Although the FUS-LC sequence is quasi-repetitive, sequences longer than four amino acids (e.g., SGYSQ, YSSYG, etc.) generally occur only once. Thus, the specific sequence in residues 39–95 may form a cross-β structure with somewhat greater stability than other segments of FUS-LC, perhaps due to more favorable arrangements and interactions among sidechains within the fibril core. Alternatively, subtle sequence differences may affect the entropic cost or desolvation energy for converting dynamically disordered segments to cross-β structures (Thirumalai et al., 2012). A core that is formed by a single contiguous portion of the FUS-LC sequence may also be intrinsically more stable than a core that is formed by segments that are separated by long loops, because the latter case requires both ends of the loops to have fixed positions, thereby increasing the total entropic cost of polymerization (Zhou, 2004).
Three Tyr sidechains (Y50, Y66, and Y75) are buried in the fibril core, while others are solvent-exposed or may interact weakly with segments of FUS-LC that do not have detectable or assignable solid state NMR signals. Although sidechain conformations are undetermined, Y50 and Y66 may engage in π-stacking interactions that help stabilize the core structure. The sidechain of D46, the only charged residue in the core-forming segment, is either solvent-exposed or interacts with sidechains of S44 and Q52, thereby reducing electrostatic repulsions by increasing the local effective dielectric constant. Buried sidechains of Q69, Q73, Q88, and Q93 may participate in polar zipper interactions along the fibril growth axis (Perutz et al., 1994).
Sidechains of certain polar residues may participate in inter-residue hydrogen bonds that help stabilize the core. For example, Y75, T78, and S84 may form a hydrogen bond network that stabilizes the loop between residues 74 and 87. Folding of residues 44–50 against residues 64–80 may be stabilized by interactions of S48 with Q69 and T71 and interactions of T45 and T47 with S77. Additionally, S70, Q73, S90, Q93, and S95 may interact with one another to stabilize folding of residues 69–73 against residues 90–95.
What distinguishes FUS-LC fibrils from pathogenic amyloids?
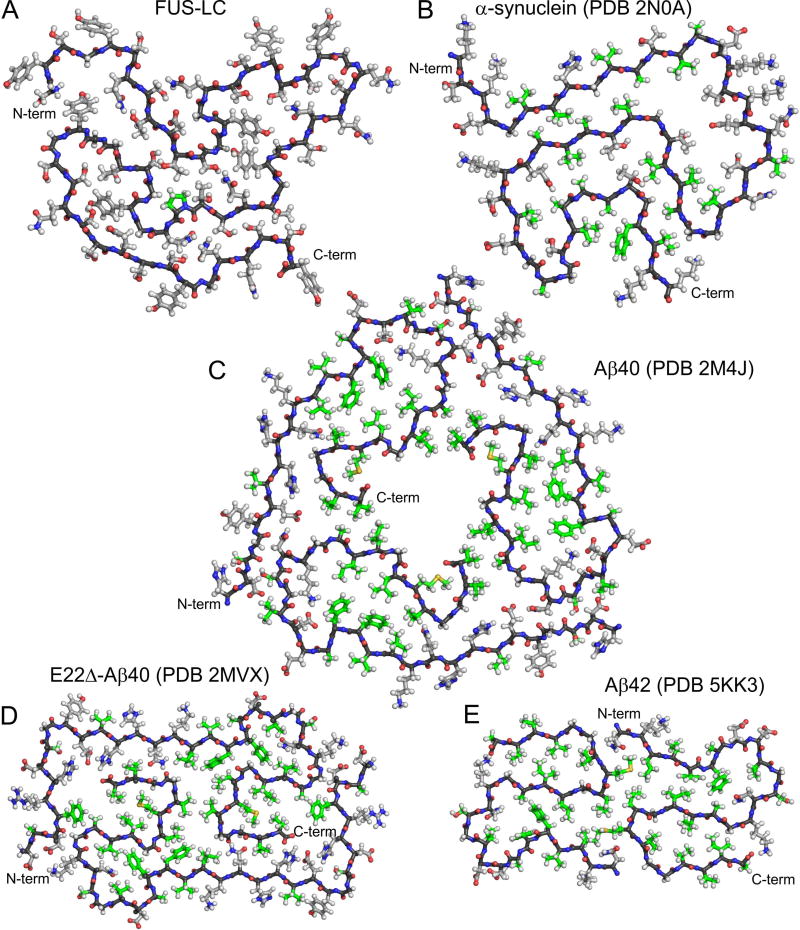
Detailed molecular structural models for wild-type 40- and 42-residue Aβ (Aβ40 and Aβ42) fibrils (Colvin et al., 2016; Lu et al., 2013; Walti et al., 2016; Xiao et al., 2015), Glu22-deletion-mutant Aβ40 (E22Δ-Aβ40) fibrils (Schutz et al., 2015), and α-synuclein fibrils (Tuttle et al., 2016) have been developed recently from solid state NMR data. These models are compared with our model for the FUS-LC fibril core in Fig. 7. In common with the FUS-LC fibril core, the pathogenic fibrils contain in-register parallel cross-β structures and have backbone conformations comprised of β-strand segments of varying lengths (from two to nine residues), irregular bends, and loops. While FUS-LC and α-synuclein fibrils indicate a single cross-β unit within each fibril, Aβ fibrils contain two or three cross-β units, arranged with twofold or three-fold symmetry about the fibril growth axis. The backbone conformation of the FUS-LC core is remarkably similar to that of α-synuclein (Fig. 7B), despite the complete lack of sequence homology.
Fig. 7. Comparison of FUS-LC fibril structure with amyloid fibril structures developed previously from solid state NMR data.
(A) FUS-LC fibril core. (B) α-synuclein fibril core, showing residues 43–97. (C) Brain-derived Aβ40 fibril, showing residues 7–40. (D) E22Δ-Aβ40 fibril. (E) Aβ42 fibril core, showing residues 15–42. In all panels, a single molecular layer is shown, viewed down the fibril growth axis. Backbone carbon atoms are black. Sidechain carbon atoms of hydrophobic residues (Ala, Val., Ile, Leu, Pro, Met, and Phe) are green. Other sidechain carbon atoms are grey.
The most striking difference between FUS-LC fibrils and pathogenic amyloid fibrils is the nature of sidechain-sidechain interactions in the fibril cores. Both α-synuclein and Aβ fibrils are stabilized by multiple intramolecular and intermolecular hydrophobic interactions in their cores, involving sidechains of Ala, Val, Leu, Ile, Met, and Phe residues (green in Fig. 7). These hydrophobic interactions augment the intermolecular backbone hydrogen bonding interactions of the cross-β architecture. The absence of purely hydrophobic sidechains (except Pro72) in the core of FUS-LC fibrils likely contributes to their lability to disassembly (Kato et al., 2012).
The FUS-LC fibril core has a higher density of polar sidechains, which may participate in hydrogen bonding networks as discussed above. Cross-β crystal structures of Ser-containing peptides determined by Eisenberg and coworkers (Sawaya et al., 2007) provide relevant examples of these networks. In the case of SSTSAA peptide crystals (PDB 20NW), in-register parallel β-sheets stack on one another such that Ser1 sidechains of one sheet interdigitate with Ser2 and Ser4 sidechains of an adjoining sheet, with 2.8 Å inter-sheet oxygen-oxygen distances, consistent with direct inter-sheet hydrogen bonds among Ser sidechains. Additionally, single-file rows of water molecules bind between Thr3 sidechains in one sheet and S2 sidechains of an adjoining sheet, with 2.8 Å distances between water oxygens and sidechain hydroxyl oxygens of both Thr3 and Ser2, consistent with water-mediated hydrogen bonding. Similar direct and water-mediated hydrogen bonds, as well as electrostatic dipole-dipole interactions among polar sidechains, may contribute to the stability of the FUS-LC core structure.
The abundance of hydroxyl-containing sidechains in the FUS-LC core provides a mechanism by which the thermodynamic stability of FUS-LC polymers could potentially be modulated by phosphorylation. In the case of purely pathogenic fibrils, the relative scarcity of hydroxyl-containing sidechains in the core may reflect the absence of evolved functions that facilitate regulation of polymerization.
The non-polymorphic character of FUS-LC fibrils distinguishes them from Aβ and α-synuclein fibrils, but is reminiscent of biologically functional fibrils formed by the fungal HET-s prion protein (Van Melckebeke et al., 2010). We offer the speculative hypothesis that non-polymorphic polymers, such as those formed by the HET-s and FUS-LC polypeptides, may represent the proper biologic function of these gene products. Proteins that assemble into a variety of polymorphic conformations, such as Aβ and α-synuclein, may instead represent aberrant assemblies unrelated to biologic function. The absence of hydrophobic interactions in the fibril core may allow self-assembling FUS-LC chains to anneal their structures efficiently to the global free energy minimum, rather than being trapped in local minima that represent polymorphs.
Phosphorylation-mediated inhibition of FUS-LC polymerization and phase separation
Proteomic analysis of FUS-LC following exposure to DNA-PK and ATP identified 14 phosphorylated residues, including six localized between residues 37 and 95 (Fig. 5). DNA-PK-mediated phosphorylation of the LC domain has previously been reported to inhibit binding of soluble GFP:FUS-LC to mCherry:FUS hydrogels (Han et al., 2012). Phosphorylation by DNA-PK also causes FUS-LC droplets to dissolve, suggesting the involvement of similar intermolecular interactions in the phenomena of LC polymerization and LC phase separation. To investigate the site specificity of phosphorylation effects, we systematically mutated pairs Ser and Thr residues to Ala, thereby blocking phosphorylation at the mutated sites. Data in Fig. 6 show strong dependences on the locations of Ala substitutions, for both hydrogel binding and droplet melting. The strongest effects are observed when phosphorylation sites in residues 42–87 are eliminated (Figs. 6B, C and E). Elimination of phosphorylation sites outside the fibril core-forming region identified by solid state NMR generally has weaker effects. These results argue in favor of a role for molecular structure in both polymerization and phase separation by FUS-LC, and especially for the relevance of the fibril core structure in Fig. 4 to these two phenomena. Alternative interpretations of the data in Fig. 6 based purely on variations in the amino acid composition or in the density of phosphorylation sites along the FUS-LC sequence would not explain, for example, why Ala substitutions at residues 61 and 68 or residues 42 and 84 have large effects, while Ala substitutions at residues 7 and 11 or residues 112 and 117 have small effects.
We suggest that phosphorylation of Ser and Thr residues within the fibril core-forming segment inhibits hydrogel binding and droplet formation most strongly because phosphorylation at these sites interferes with cross-β interactions. If assembled as in-register, cross-β polymers similar to the structural model in Fig. 4, core-forming residues of FUS-LC would be separated by about 4.7 Å. Negative charges introduced by phosphorylation would be expected to exert repulsive forces that reduce polymer stability.
Our results from experiments involving FUS-LC phosphorylation are consistent with previous studies of the LC domain of hnRNPA2. Correlative mutagenesis and chemical footprinting studies have given evidence that the same structures and forces account for the ability of the hnRNPA2 LC domain to partition into either hydrogels or liquid-like droplets (Xiang et al., 2015). Likewise, the correlative effects of aliphatic alcohols on the melting of both hydrogels and liquid-like droplets indicate that these two states of phase separation are driven by related chemical forces (Lin et al., 2016). If correct, these studies, coupled with the data presented herein, offer a satisfying unity to studies on LC domains as phase-separated into either liquid-like droplets or hydrogels.
We close with a point emphasized repeatedly in previous studies (Kato et al., 2012; Kwon et al., 2013; Lin et al., 2016; Shi et al., 2017; Xiang et al., 2015). LC domain polymers characterized in this and earlier reports, when employed in living cells and subject to many forms of post-translational modification, are undoubtedly far more dynamic than the long amyloid-like fibrils that are formed by self-assembly of unmodified polypeptides purified in recombinant form from bacteria. We hope that studies of DNA-PK-mediated phosphorylation of the FUS-LC domain may offer an initial glimpse of the rich and complex web of cellular processes involved in regulating the dynamics of LC domain polymerization.
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Robert Tycko (robertty@mail.nih.gov).
EXPERIMENTAL METHOD AND SUBJECT DETAILS
Proteins were expressed at 37° C in BL21(DE3) or BL21(DE3)pLysS E. coli cells, using M9 medium for isotopically labeled proteins and Luria-Bertani (LB) broth for unlabeled proteins.
METHOD DETAILS
Production of FUS-LC proteins
Unless otherwise specified, all chemicals were purchased from Sigma-Aldrich. Recombinant U-FUS-LC was expressed in BL21(DE3)pLysS E. coli cells (Invitrogen). 50 ml of overnight culture was added to 1 L of M9 salts (or LB broth for unlabeled samples) and cells grown to an optical density of OD600 = ~1.0 in an incubator/shaker at 37 °C and 220 revolutions per minute (rpm). Protein expression was induced by adding 1 ml of 0.5 M IPTG to the culture and growing for another 1.5 h at 37 °C and 220 rpm. Typically, about 6 g of wet cells were harvested by centrifugation from 2 L of culture. The pellet was flash frozen in liquid nitrogen and stored at −80 °C until purification. Cells were thawed on ice for 15 min, then suspended in 40 ml of 50 mM Tris-HCl (pH 7.5), 500 mM sodium chloride, 1% v/v Triton X-100, 6 M guanidinium hydrochloride, with two tablets of Roche cOmplete EDTA-free protease inhibitor. Lysozyme powder was added to a final concentration of 1 mg/ml and 8 µl of Pierce Universal Nuclease for Cell Lysis (Thermo Fisher Scientific) was added. The cell suspension was sonicated in an ice bath for 10 min at 0.4 output, 30% duty cycle, using a Branson Sonifier 250 before rotating at 4° C for 10 min. The mixture was centrifuged for 30 min at 223,000 X g and 4°C to produce a clarified lysate. The supernatant was added to 5 ml of Qiagen Ni-NTA agarose resin equilibrated in 20 mM sodium phosphate, 500 mM sodium chloride, and 8 M urea, pH 7.4, and rotated at 4° C for 30 min. The slurry was poured over a 1.5 × 20 cm gravity flow column (Bio-Rad Laboratories), washed with about 200 ml of 20 mM sodium phosphate, 500 mM sodium chloride, 8 M urea, and 20 mM imidazole, pH 7.4 until the optical absorbance of the flow-through at 280 nm was less than 0.15 (1 cm path length). His-tagged FUS-LC was eluted using 20 mM sodium phosphate, 500 mM sodium chloride, 8 M urea, and 200 mM imidazole, pH 7.4. Typically, 10–15 mg of protein was obtained from 2 L of culture. 1 ml aliquots of the purified protein (2–3 mg/ml) were prepared, flash frozen in liquid nitrogen and stored at −25 °C.
For hydrogel binding, mCherry- and GFP-FUS-LC were prepared as described previously (Kato et al., 2012). For droplet formation experiments, the last proline residue in the His6-tag linker region of His6-tagged FUS-LC was mutated to lysine (His6-K-FUS-LC) using the QuikChange Site-Directed Mutagenesis kit (Agilent, Santa Clara, CA). GFP- and His6-K-FUS-LC double mutants were generated using the QuikChange Multi-Site-Directed Mutagenesis kit (Agilent, Santa Clara, CA). These mutants were overexpressed and purified as described above. DNA-PK was purchased from Promega (Madison, WI).
Production of segmentally labeled FUS-LC
Segmentally-labeled FUS-LC was prepared with intein protein ligation technology. We used fused split intein AvaDnaE (Shah et al., 2012), kindly provided by Prof. Tom W. Muir of Princeton University. The design of the N-terminal and C-terminal fragments is summarized in Figure S4A. All DNA fragments were amplified by either a one-step or a two-step PCR technique and sub-cloned in pHis-parallel vector (Sheffield et al., 1999). The expression plasmids were transformed in BL21(DE3) E.coli strain. Overexpression of non-labeled fragments was carried out in LB medium as described previously (Kato et al., 2012). 15N,13C- labeled fragments were overexpressed in M9 medium with 13C-glucose and 15N-ammonium chloride as carbon and nitrogen sources. Briefly, a single colony of BL21(DE3) cells was inoculated in 10 ml of LB media and incubated at 37° C overnight without shaking. The pre-culture (5 ml) was transferred into 1 L of M9 medium containing isotopically labeled compounds and shaken at 37° C until OD600 reached 0.6–0.8 (6–7 h incubation). The culture temperature was decreased to 20° C and IPTG was added to the M9 medium to a final concentration of 0.5 mM. Incubation was continued overnight. Cells were harvested by centrifugation, washed with PBS, then frozen and stored at −80° C.
All fragments were purified in semi-denaturing conditions as follows. Cells were resupended in lysis buffer containing 50 mM Tris-HCl pH 8.0, 500 mM NaCl, 1 mM TCEP, 1% Triton X-100, 2 M urea, and protease inhibitor cocktail. Lysozyme (100 mg/ml) was added to the cell suspension to a final concentration of 0.2 mg/ml. After incubation on ice for 30 min, cells were sonicated for 2 min (cycle of 10 s on and 30 s off at 65% power, Fisher Scientific Model FB705) and clarified by ultracentrifugation at 35,000 rpm for 1 h at 4° C. The supernatant was mixed with Ni-NTA resin (Qiagen, USA), which was pre-equilibrated with wash buffer containing 20 mM Tris-HCl, pH 7.5, 500 mM NaCl, 1 mM TCEP, 0.1 mM PMSF, and 2 M urea. The mixture was gently shaken in a cold room for 30 min, then poured into a glass column. The Ni-NTA resin was washed with wash buffer with 20 mM imidazole. Bound proteins were eluted with wash buffer with 250 mM imidazole. Eluted proteins were concentrated with Amicon Ultra centrifugal filters (Millipore, USA) to the final protein concentration of 2–3 mM. Concentrated proteins were centrifuged to remove any particles, aliquoted in microcentrifuge tubes, and stored at −80° C.
Intein reactions (5–10 ml) were carried out in a mixture containing 200 µM of the N-terminal fragment and 200 µM of the C-terminal fragment in 40 mM potassium phosphate, pH 7.2, 150 mM NaCl, 1 mM TCEP, 400 mM sodium 2-mercaptoethanesulfonate (MES Na), and 2 µg/ml Caspase-3. The mixture was gently rotated at room temperature for 48 h, then incubated on ice for 1 h. The mixture was centrifuged at 4000 rpm for 30 min at 4° C. The product (full-length FUS-LC) was only present in the pellet. To purify the product, the pellet was dissolved in post-reaction purification (PRS) buffer 1 (5 ml) containing 20 mM Tris-HCl, 200 mM NaCl, 20 mM EDTA, 0.1 mM PMSF, and 2 M Gdn-HCl. The protein solution was loaded on a Ni-NTA column (5 ml). The column was washed with 50 ml Ni-NTA buffer supplemented with 20 mM imidazole. The bound proteins were eluted with Ni-NTA buffer with 250 mM imidazole. Eluted proteins were concentrated with Amicon Ultra filters to a 2 ml volume and loaded on a gel filtration column (Superdex S-200, GE Health Science, USA) that was equilibrated with PRS buffer 2 containing 20 mM Tris-HCl, 200 mM NaCl, 5 mM DTT, 0.5 mM EDTA, 0.1 mM PMSF, and 2 M Gdn-HCl. Fractions were analyzed by SDS-PAGE, combined for the peak of the product, and concentrated to the final protein concentration of 20–30 mg/ml.
Fibril formation conditions
FUS-LC fibrils were prepared by concentrating purified FUS-LC to 30 µM in about 100 µl final volume, using a 0.5 ml, 10 kDa, centrifugal filter (Millipore), then diluting to 500 µl with 20 mM sodium phosphate, 200 mM sodium chloride, 20 mM β-mercaptoethanol, 0.1 mM phenylmethylsulfonyl fluoride, pH 7.4, and concentrating again to about 100 µl volume. The dilution and concentration steps were repeated three times. The sample was then incubated quiescently at 24° C for six days. Fibril formation was verified by TEM. To prepare seeds, fibrils were sonicated with a Branson Sonifier 250 at 0.15 output, 10% duty cycle for 10 min, yielding fragments with lengths of 50–100 nm. Seeded fibril growth was performed by adding the seeds to 100 µM monomeric FUS-LC, which had been dialyzed extensively against 20 mM sodium phosphate, pH 7.4 to remove urea and other components of the purified aliquots described above. The molar ratio of monomeric FUS-LC to FUS-LC in the seeds was 19:1. Long fibrils were observed by TEM after 3–7 d. The fibrils were then sonicated again to create additional seeds, which were frozen in liquid nitrogen in 100 µl aliquots and stored at −20° C. U-FUS-LC fibrils for NMR experiments were prepared by dialyzing purified U-FUS-LC against 20 mM sodium phosphate buffer, pH 7.4 at a concentration of 40–80 µM. Aliquots of frozen seeds were added, to a final 19:1 molar ratio of soluble protein to protein in seeds. The sample was then incubated quiescently at room temperature for 7 days. For NMR measurements, fibrils were then pelleted for 1–2 h at 247,000 X g and 4° C in a Beckman Optima ultracentrifuge. Fibril pellets were packed into MAS rotors (3.2 mm outer diameter) by centrifugation at 50,000 X g and 4° C for 1–20 h, using a home-made device to hold the MAS rotor and the centrifuge tube containing the fibril pellet.
For segmentally labeled samples, 250–350 µl aliquots of purified protein at 30–70 µM were diluted into 1 ml volumes with 20 mM sodium phosphate buffer, pH 7.4, 5 mM dithiothreitol. Fibril growth was initiated by adding seeds to the diluted protein solutions, in this case with a 99:1 molar ratio. Samples were incubated quiescently at room temperature for 7–11 days. Fibrils were then pelleted by ultracentrifugation for 1–2 h at 247,000 X g and 4° C Fibril pellets were packed into MAS rotors (1.8 mm outer diameter) by centrifugation at 17,000 X g and 24° C for 5–20 mm.
Electron microscopy
TEM images were obtained on a FEI Morgagni microscope, equipped with side-mounted Advantage HR and bottom-mounted XR-550B cameras (Advanced Microscopy Techniques) and operating at 80 keV. Negatively-stained samples were prepared on home-made 3–6 nm carbon films, supported by lacey carbon on 300 mesh copper grids (Electron Microscopy Sciences). Grids were glow-discharged immediately before sample application. A 5 µl aliquot of unpelleted fibril solution, typically diluted by a factor of 20, was adsorbed to the carbon film for two minutes, blotted, washed with 5 µl of water for 10 s, blotted, washed again, blotted, stained with 5 µl of 3% uranyl acetate for 10 s, blotted, and dried in air. Images were recorded with the side-mounted camera.
For MPL measurements, a 5 µl aliquot of unpelleted fibril solution was adsorbed to a freshly glow-discharged grid, along with an additional 5 µl aliquot of dilute TMV (generously provided by the laboratory of Prof. Gerald Stubbs, Vanderbilt University). After 5 min of incubation, the grid was washed as described above, but not stained. Dark-field TEM images were recorded with the bottom-mounted camera, using a 1.2° tilt of the electron beam, optimization of the beam for uniform illumination of the field of view, and minimization of the exposure time as previously described (Chen et al., 2009). Images were analyzed with ImageJ software (available from https://imagej.nih.gov/ij).
MPL counts in Fig. 1E (units of kDa/nm) were determined by the equation:
where IF is the integrated image intensity within a 80 nm X 63 nm rectangle centered on a FUS-LC fibril segment, and IB1 and IB2 are the integrated background intensities within identical rectangles on both sides of the fibril (Fig. S1C). ITMV is the average of integrated intensities for many 80 nm X 63 nm rectangles centered on TMV particles, after background subtraction, and 131 kDa/nm is the standard MPL value for TMV. A total of 123 MPL counts were obtained from three dark-field images, with 9–23 TMV intensities per image.
MPL error counts in Fig. S1D (units of kDa/nm) were determined by the equation:
where IB is the integrated background intensity within a single 80 nm X 63 nm rectangle, IAVE is the average of all background intensities, and Itmv has the same definition as above. Gaussian fitting of MPL and error histograms and data plotting were performed using Igor Pro 6.3.7.2 (WaveMetrics).
Atomic force microscopy
Fibrils for AFM measurements were prepared by diluting a 100 µl aliquot of 0.1 mM FUS-LC fibrils with 300 µl of 20 mM sodium phosphate buffer, pH 7.4. The fibrils were sonicated with a Branson 250 sonifier using a small horn attachment with a 10% duty cycle, 0.15 output setting for 10 min to create seeds. A 1.0 ml aliquot of unassembled FUS-LC at a concentration of 44 µM was prepared by extensive dialysis against 20 mM sodium phosphate, pH 7.4, added to 80 µl of the fibril seed solution, mixed gently with a pipette, and incubated quiescently at room temperature for four days. AFM samples were prepared by diluting 25 µl of the fibril solution into 950 µl of 20 mM sodium phosphate, pH 7.4, with gentle vortexing. A 50 µl aliquot was then applied to a freshly cleaved mica surface, incubated for 2 min, blotted with tissue paper, and dried under a gentle stream of nitrogen gas. The surface was imaged on a Veeco Multimode AFM instrument with Nanoscope IV controller, operating in tapping mode with a scan rate of 0.8 Hz, acquiring 256 points per line and 256 lines over a 9 µm2 area. The cantilever was n-type Si with dimensions 125 µm × 45 µm × 0.4 µm with a tip radius 5–6 nm and 40 N/m force constant (model ACT-20, Allied Nanostructures, Mountain View, CA), oscillating at a frequency of approximately 300 kHz. Fibril diameters were measured by subtracting the average baseline in a 1 µm cross-section from the fibril peak height. The value of 5.5 ± 0.7 nm (average ± standard deviation) is the result of 21 measurements on eight different fibrils in four different images.
Thioflavin T and Congo Red binding assays
FUS-LC fibrils were prepared by adding 40 µl of a 25 µM solution of seeds to 500 µl of FUS-LC solution at 40 µM in 20 mM sodium phosphate buffer, pH 7.4. Fibril growth proceeded for 24 h. A stock solution of thioflavin T (ThT) was prepared by dissolving 7.2 mg of ThT in 1.44 ml of 20 mM sodium phosphate buffer, pH 7.4, and diluted with phosphate buffer to 20 µM ThT for fluorescence measurements. A 10 µl aliquot of FUS-LC fibrils (or unseeded, soluble FUS-LC at 40 µM concentration) was added to 200 µl of 20 µM ThT, vortexed briefly, then loaded into the cuvette of a StellarNet BLACK-Comet-TEC fluorescence spectrometer, operating with excitation at 423 nm from a monochromatic light-emitting diode. Fluorescence emission spectra were averaged over four scans, with 500 ms acquisition per scan. Enhanced ThT fluorescence at 500 nm is a standard indication of ThT binding to cross-β structures.
For Congo Red (CR) binding, a stock solution of 1 mM CR was prepared in 20 mM sodium phosphate buffer, pH 7.4, and diluted with phosphate buffer to 6 µM CR for absorbance measurements. For each measurement, 125 µl of FUS-LC (in fibrillar or soluble states) was added to 125 µl of 6 µM CR, vortexed briefly, then incubated for 10 min at room temperature. Absorbance spectra were recorded on a TECAN Infinite M200Pro plate reader. Aliquots of 200 µl were loaded into a polystyrene clear-bottom, chimney-well, 96-well plate (Geriner BioOne) for each measurement. Enhanced and red-shifted CR absorbance are standard indications of CR binding to cross-β structures.
Predictions of amyloid formation
All predictions were run on residues 2–214 of the FUS-LC sequence, using the web servers provided by the authors of the prediction programs, which are available at http://bioinf.uab.es/aggrescan/ (for Aggrescan), http://www.comprec.pwr.wroc.pl/fish/fish.php (for FISH), http://bioinfo.protres.ru/fold-amyloid (for FoldAmyloid), http://services.mbi.ucla.edu/zipperdb (for ZipperDB), http://metamyl.genouest.org/e107_plugins/metamyl_aggregation/db_prediction_meta.php (for MetAmyl), http://protein.bio.unipd.it/pasta2 (for PASTA 2.0), http://waltz.switchlab.org (for WALTZ), and http://www-mvsoftware.ch.cam.ac.uk/index.php/zyggregator (for Zyggregator). FISH was run with default parameters (i.e., threshold = 0.19, prediction = "standard"). FoldAmyloid was also run with default parameters (i.e., scale = "Expected number of contacts 8 Å", averaging frame = 5, and threshold = 21.4). MetAmyl was run with its threshold set to "best global accuracy". PASTA 2.0 was run with "top pairing energies" = 20 and "energy threshold" = −5. WALTZ was run at pH 7, with its threshold set for "best overall performance". Zyggregator was run at pH 8. Residue numbers in Fig. S7 represent either the first or the central residue in a short peptide segment, depending on the details of the prediction algorithm.
NMR measurements
Measurement conditions, including sample quantities, pulse sequence parameters, and total measurement times, are given in Table S3. Processed 2D and 3D spectra are available in Sparky format at http://dx.doi.org/10.17632/rxbh442x2g.1.
NMR measurements were performed at 21.1 T, 17.5 T, 14.1 T, and 9.4 T magnetic field strengths. Data at 21.1 T (895.1 MHz 1H NMR frequency) were obtained at the National High Magnetic Field Laboratory (NHMFL, Tallahassee, FL), using a Bruker Avance III spectrometer console and a low-E MAS NMR probe produced by Peter Gor'kov of NHMFL, with 3.2 mm MAS rotors. Data at 17.5 T (746.1 MHz 1H NMR frequency) were obtained with a Varian Infinity console and either a 3.2 mm Bruker E-Free probe, a 1.8 mm MAS NMR probe produced by the laboratory of Dr. Ago Samoson (Tallinn University of Technology, Estonia), or a 3.2 mm low-E MAS probe produced by Black Fox, Inc. (Tallahassee, FL). Data at 14.1 T (599.2 MHz 1H NMR frequency) were obtained with a Varian InfinityPlus console and either a 3.2 mm Varian BioMAS probe or a 1.8 mm MAS NMR probe produced by the laboratory of Dr. Samoson. Data at 9.4 T (400.6 MHz 1H NMR frequency) were obtained with a Bruker Avance III console and a 3.2 mm Varian T3 MAS NMR probe. Sample temperatures were maintained at approximately 20° C in all experiments. 13C-13C polarization transfers in 2D and 3D spectra used Dipolar-Assisted Rotational Resonance (DARR) (Takegoshi et al., 2001). 15N-13C polarization transfers used CP (Pines et al., 1973). 1D data were processed with Varian Spinsight or Bruker TopSpin software. 2D and 3D data were processing with NMRPipe software (Delaglio et al., 1995) and analyzed with Sparky software (https://www.cgl.ucsf.edu/home/sparky). Pure Gaussian apodization functions were used to process all data, with no artificial resolution enhancement to reduce apparent NMR linewidths.
Quantitative measurements of 13C-13C and 15N-15N dipole-dipole couplings were performed with the PITHIRDS-CT and 15N-BARE techniques, respectively, as previously described (Hu et al., 2012; Tycko, 2007). PITHIRDS-CT measurements were performed on fully hydrated FUS-LC fibril samples in which backbone carbonyl sites of either all Tyr residues or all Thr residues were 13C-labeled by overexpression with M9 medium that contained all amino acids, including either 1-13C-Tyr or 1-13C-Thr. PITHIRDS-CT and 15N-BARE data were analyzed by comparison with numerical simulations, as previously described (Hu et al., 2012; Tycko, 2007). A 10% signal contribution from natural-abundance carbonyl 13C, estimated by assuming that the PITHIRDS-CT signals include contributions from five labeled residues and 50 unlabeled residues and that natural-abundance 13C signals from unlabeled residues are independent of the dipolar evolution period, was subtracted from PITHIRDS-CT data in Fig. S6B.
MCASSIGN calculations
Site-specific chemical shift assignments for the structured core of FUS-LC fibrils (Table S1) were obtained from two independent sets of MCASSIGN calculations (Bayro et al., 2014; Hu et al., 2011). In the first set of calculations, crosspeaks in 3D NCACX, NCOCX, and CONCA spectra were picked manually in Sparky. Crosspeak lists from Sparky were converted into NCACX, NCOCX, and CONCA signal tables with the MCASSIGN format, with 55, 49, and 57 signal rows, respectively. Chemical shift uncertainty values estimated from signal-to-noise ratios and linewidths (0.25–0.40 ppm for most signals, 0.50–0.85 ppm for weaker signals).
In preliminary calculations (using the mcassign2b version of the MCASSIGN program), no signals were assigned consistently to the C-terminal half of the FUS-LC sequence. Subsequent calculations therefore used only residues 1–112. Weighting factors in the MCASSIGN assignment score for good connections, bad connections, assignment edges, and used signals (Bayro et al., 2014; Hu et al., 2011) were incremented from 0 to 10, 10 to 60, 0 to 2, and 0 to 2, respectively, in 35 annealing steps, with 1 X 108 Monte Carlo attempts in each step. The acceptance fraction threshold value was set to 0.001, with final incrementation of the weighting factors in the final five annealing steps. In a first round of 50 independent mcassign2b runs, identical signals from all three signal tables were assigned 50 times to residues 44–47, 49–52, 63–66, and 74–84. These definite assignments were added to the signal tables, and chemical shift uncertainties for the remaining unassigned signals were increased by 0.1 ppm.
A second round of 50 independent mcassign2b runs was then performed, using the same parameters, but with 20 annealing steps. Additional signals from at least one of the three signal tables were assigned identically in all 50 runs to residues 48, 85, 87, and 89, and 94, and in at least 39–48 out of 50 runs to residues 86, 88, 89, and 94. The signal tables were then updated to include these additional definite assignments. 2D 15N-13C NMR spectra of N60- and C60-FUS-LC fibrils (Fig. S5F,G) were also used to confirm tentative assignments for residues 40 and 92, based on the observation that a 15N-13Cα crosspeak, which was assigned in all three signal tables to G40 in 40 out of 50 mcassign2b runs, appears in the 2D spectrum of N60-FUS-LC fibrils, and the observation that a 15N-13Cα crosspeak, which was assigned in all three signal tables to G92 in 49 out of 50 mcassign2b runs, appears in the 2D spectrum of C60-FUS-LC fibrils.
A third round of 50 independent mcassign2b runs was then performed, with the same parameters as the second set. In the third set of runs, additional signals in all three signal tables were assigned identically in all 50 runs to residues 39, 72, 86, 88, 90–91, and 93–95, and in two of the three signal tables for Q73. Tentative assignments for residues T68 and Q69 were also confirmed, based on the absence of the corresponding signals from the 2D 15N-13C NMR spectrum of N60-FUS-LC fibrils and the presence of these signals in the 2D spectrum of C60-FUS-LC fibrils. Signal tables were then updated to include the additional definite assignments, and a final round of 50 independent mcassign2b runs was performed, using 20 annealing steps and 106 attempts in each step. Definite signal assignments in all three signal tables were obtained for S70, and in the remaining signal table for Q73. Signals for G67 were assigned in 47 out of 50 runs. Final results from this set of MCASSIGN calculations are summarized in Table S1 and Fig. S3. Chemical shifts were deposited in the Biological Magnetic Resonance Data Bank with accession code 30304.
As an additional check on the accuracy of the assignments in Table S1, a second set of MCASSIGN calculations was performed, using a different approach. These calculations used a modified program, called mcassign2c, in which the definitions of good and bad connections were modified so that each connection (i.e., each comparison of assigned chemical shifts from different signal tables, with chemical shift values δ1 and δ2 and uncertainties ε1 and ε2) contributed a quantity xg to the total number of good connections and a quantity Xb to the total number of bad connections, given by
and xb = 1 - xg with . With these definitions, a given connection is considered entirely good if the chemical shift difference is less than or equal to , or entirely bad if the chemical shift difference is greater than or equal to , and changes linearly from good to bad within these limits. The value of q was set to 3.0. (In mcassign2b, each connection is either entirely good or entirely bad, equivalent to q = 1.0.)
For the second set of MCASSIGN calculations, 3D NCACX, NCOCX, and CONCA spectra of U-FUS-LC fibrils were reanalyzed in Sparky, leading to signal tables with 64, 50, and 58 signal rows, respectively. In addition, signals from the 3D CANCX spectrum of U-FUS-LC fibrils were divided into two signal tables, with 26 and 23 signal rows, differing in whether residue-type assignments for each row were based on 13Cα chemical shifts in the t1 dimension or 13Cα chemical shifts in the t3 dimension. The full FUS-LC sequence was considered. 3D NCACX and CONCA spectra of N112-FUS-LC fibrils were analyzed and compared with the corresponding 3D spectra of U-FUS-LC fibrils, allowing assignments of 22 NCACX signals and 33 CONCA signals to be restricted to residues 1–112 (represented by upper-case letters in the mcassign2c input files) and excluded from residues 113–214 (represented by lower-case letters).
A first round of 20 independent mcassign2c runs was performed, with 40 annealing steps and 2 X 108 Monte Carlo attempts in each step. Weighting factors for good connections, bad connections, and assignment edges were incremented from 0 to 4, 0 to 12, and 0 to 0.5, respectively, with no weighting of used signals. The acceptance fraction threshold value was set to 0.05, with final incrementation of the weighting factors in the final ten annealing steps. Definite assignments (i.e., identical assignments in all 20 runs) were obtained for residues 45–52, 73–77, and 80–95. After updating the signal tables to include these definite assignments, two additional rounds of mcassign2c runs was performed, with updates to the signal tables after each round. After the third round, in which the number of attempts per annealing step was increased to 8 X 108, definite assignments were obtained for residues 44–54, 68–70, and 72–95. No signals were assigned consistently to residues 113–214.
At this point, the 3D spectra were re-examined and weak signals were eliminated from the MCASSIGN signal tables, leaving 49, 43, and 44 signal rows in the NCACX, NCOCX, and CONCA signal tables, respectively, and 21 and 23 signal rows in the two CANCX signal tables. A final round of 20 independent mcassign2c runs was performed, using only residues 1–112 of the FUS-LC sequence and with 1 X 108 attempts per annealing step. Again, definite assignments were obtained for residues 44–54 and 68–70, and 72–95.
Assignments from the second set of MCASSIGN calculations are in full agreement with Table S1. Additional assignments in Table S1 for residues 39–40 and 63–67 are primarily a consequence of including information from 2D spectra of N60-FUS-LC fibrils and C60-FUS-LC fibrils in the first set of MCASSIGN calculations.
From the original NCACX, NCOCX, and CONCA signal tables in the second set of MCASSIGN calculations (including weak signals that were later eliminated), 28, 18, and 21 signal rows remain unassigned, respectively. From the two CANCX tables, 12 and 3 signal rows remain unassigned. One possible explanation for these unassigned signals is that they arise from a minor polymorph in the FUS-LC fibril samples, with a distinct structure and a distinct set of solid state NMR signals. To test this possibility, mcassign2c calculations were run with signal tables that included only the remaining unassigned signals. These calculations yielded no consistent assignments, arguing against the existence of a minor polymorph. Instead, as discussed in the main text, unassigned signals are attributable to partially ordered residues or segments of the FUS-LC sequence that are outside of residues 39–95.
Calculation of structural models for the FUS-LC fibril core
Structure calculations were performed using the Xplor-NIH package on the NIH Biowulf high-performance computing cluster. Nine copies of FUS-LC residues 37–97 were initially prepared in extended conformations, with a 7 Å spacing between monomers and with the peptide chains perpendicular to the z-axis (fibril growth axis). In the first round of calculations, 448 independent Xplor-NIH runs were performed, using 5000 steps of torsion angle dynamics at 4000 K followed by annealing to 10 K in 10 K decrements with 3000 steps of torsion angle dynamics at each temperature and then by final energy minimizations in torsion angle and Cartesian coordinates. Non-crystallographic symmetry and translational symmetry potentials (Xplor-NIH PosDiffPot and DistSymmPot potentials) were used to keep the conformations and orientations of the nine monomers identical along the fibril growth axis. Intermolecular distance restraints (Xplor-NIH NOE potentials) were applied between all neighboring molecules for pairs of carbonyl carbons of Thr and Tyr residues in the fibril core (residue numbers 41, 45, 47, 64, 66, 68, 78, 75, 81, 91), with a 4.75 +/− 0.05 Å carbon-carbon distance, as dictated by the PITHIRDS-CT and MPL data (Figs. 1E and S6B) and consistent with standard properties of cross-β structures. Artificial "residual dipole coupling" potentials (Xplor-NIH RDC potentials) were applied to pairs of carbonyl carbons of the same residues in the three central monomers to align the fibril growth axis with the z-axis. 15N-BARE data (Fig. S6C) were represented by 3D torsion angle potential surfaces as previously described (Xplor-NIH TorsionlnterpolPot potential). To improve the efficiency of the structure calculations, the same 15N-BARE data were also represented by nitrogen-nitrogen distance restraints, using upper and lower distance limits obtained by comparing the experimental data with simulations for all possible pairs of backbone ψ angles in a tripeptide (i.e., all possible nitrogen-nitrogen distances). Upper and lower distance limits were implemented for sites with relatively rapid 15N-BARE decays (residues 48–52, 63, 65, 67–69, 75–76, 80, 82–83, 85, and 91–92). Lower limits only were implemented for sites with slower decays (residues 39–40, 46, 74, and 77–79). Backbone torsion angles were also restrained by predictions from TALOS-N (Xplor-NIH CDIH potential), calculated from the assigned 13C/15N chemical shifts and implemented with ranges equal to twice the uncertainties reported by TALOS-N (Fig. S6A; Table S1) plus an additional ±15°. Test calculations were run with only potentials derived from N-BARE data and TALOS-N predictions to determine whether the TALOS-N predictions were consistent with the 15N-BARE data. Based on these calculations, the TALOS-N predictions for the φ angles of residues 47 and 85 and the ψ angles of residue 47–48, 66–67, 81, 84, and 85 were discarded. Long-range inter-residue crosspeaks from 2D and 3D spectra of 1,3-Glyc-FUS-LC and 2-Glyc-FUS-LC fibrils were represented by 89 NOE potentials upper and lower distance limits of 8.0 Å and 2.0 Å. These NOE potentials were divided into three classes: (i) unambiguous, meaning that the assignments of both sites connected by the crosspeak were unique; (ii) partially ambiguous, meaning that the assignment of one site was unique and the possible assignments of the other site were separated by two residues or less in the FUS-LC sequence; (iii) fully ambiguous, meaning that the assignments of both sites were not unique or the possible assignments of one site were separated by more than two residues in the FUS-LC sequence. Ambiguous NOE potentials were implemented with the "soft" flag. Standard potentials of Xplor-NIH (BOND, ANGLE, IMPR, RepelPot) were used to restrain bond lengths and bond angles to the expected protein geometry and to establish atomic radii. Scale factors for all potentials are given in Table S4.
In the second round of calculations, each of the 44 lowest-energy models from the first round was used as a starting point for 112 additional runs. Torsion angle dynamics were performed at 4000 K for 1000 steps, followed by annealing with 4000 steps at each temperature from 4000 K to 10 K in 10 K decrements and by final energy minimizations in torsion angle and Cartesian coordinates. Based on an analysis of the 44 starting models, an additional square-well RDC potential, centered at 20° with a span of 40°, was applied to the carbon-oxygen bonds of backbone carbonyl groups of certain residues of the central monomer, to align these carbonyl groups approximately with the fibril growth axis, as required for intermolecular hydrogen bonds in a cross-β structure. These RDC potentials were applied only to carbonyl groups of residues that were found to participate in β-strands, defined by the criterion that their ensemble-average φ/ψ torsion angles satisfy −80° >φ > −200° and 40° < ψ < 220° (residues 44–46, 52–54, 63–64, 67–70, 83–90, and 93–95.) These artificial RDC potentials are an alternative to the intermolecular distance restraints (Colvin et al., 2016; Schutz et al., 2015; Van Melckebeke et al., 2010; Walti et al., 2016) or empirical hydrogen bond potentials (Lu et al., 2013; Tuttle et al., 2016) that have been used to align backbone hydrogen bonds in previous solid state NMR studies of amyloid and prion fibrils. In the second round of calculations, the Xplor-NIH torsionDB potential was also included to favor sidechain conformations with low energies and backbone conformations within allowed regions of Ramachandran space. Other potentials were the same as in the first round, but with scale factors as shown in Table S4.
From each of the 44 sets of runs, the 11 lowest-energy structures were examined. Only structures with no violations of the nonbonded, protein geometry, RDC, CDIH, and NOE potentials were considered further. These 78 structures were then sorted by energy, including only the contributions from nonbonded, protein geometry, RDC, CDIH, NOE, and TorsionlnterpolPot potentials, and excluding contributions from PosDiffPot, DistSymmPot, and torsionDB potentials. When more than one structure came from the same set of second-round runs (i.e., same starting point), the lowest-energy structure was retained and higher-energy structures were discarded. The 20 lowest-energy structures were then deposited in the Protein Data Bank with accession code 5W3N. Structure statistics and Protein Data Bank validation scores are given in Table S5.
A total of 51 backbone torsion angle constraints from TALOS-N, 33 backbone torsion angle potentials from 15N-BARE data, 11 intermolecular distance restraints from the PITHIRDS-CT data, and 89 long-range distance restraints from the 2D and 3D spectra were used per molecule in the structure calculations. An additional 338 inter-residue crosspeaks, connecting sites that differ in residue number by one or two, were detected but not used in structure calculations. These crosspeaks confirm the chemical shift assignments in Table S1.
Molecular dynamics simulations
Simulations were performed with NAMD (Phillips et al., 2005), using Charmm potentials, and were visualized with VMD (Humphrey et al., 1996). For Movie S1, an initial structure for nine copies of full-length FUS-LC was generated by attaching N- and C-terminal segments to the lowest-energy final structure for the FUS-LC fibril core described above, using Xplor-NIH to create and attach these segments in initially extended conformations and then to randomize their conformations while keeping the core structure fixed. NAMD simulations were then performed in vacuum, using a dielectric constant of 100 and a scaling of non-bonded interactions by 0.5 to facilitate conformational transitions of the N- and C-terminal segments. All atoms in residues 39–95 were restrained to their initial positions by harmonic potentials. Thus, these simulations were not intended to produce an accurate depiction of the dynamics of full-length FUS-LC in its fibrillar state in an aqueous environment. Rather, these simulations were intended to produce an "artist's rendition" of the relatively rigid FUS-LC core, surrounded by highly flexible N- and C-terminal segments. After an initial simulation at 127° C for 30 ns, a final simulation was performed at 37° C.
For Movie S2, the five central copies of FUS-LC residues 37–97 were taken from the lowest-energy structure described above and solvated in a 80.5 Å X 70.7 Å X 56.5 Å water box with 100 mM NaCl, using the relevant commands in VMD. NAMD simulations were performed for 150 ns at 37° C with Langevin dynamics, Nose-Hoover pressure control, and periodic boundary conditions.
Identification and quantification of FUS-LC phosphorylation sites
Phosphorylation reactions were performed in 50 µl of a reaction mixture containing 50 mM HEPES, pH 7.5, 100 mM KCl, 10 mM MgCl2, 0.2 mM EGTA, 0.1 mM EDTA, 1 mM DTT, 0.2 mM ATP, 10 µg/ml calf thymus DNA, 50 µg GFP:FUS-LC and 200 units of DNA-PK. Reaction mixtures were incubated at 30° C and quenched by addition of 150 µl of 8 M urea. For chymotrypsin digestion, phosphorylated protein samples were diluted to a final urea concentration of 2 M by addition of buffer containing 100 mM Tris, pH 8, and 10 mM CaCl2. Sequencing-grade chymotrypsin (Roche) was added to a final enzyme: substrate ratio of 1:100 (w/w). Digestion proceeded overnight at room temperature and was quenched by addition of trifluoroacetic acid to a final concentration of 0.1%. Precipitates were removed by centrifugation at 4,000 rpm for 30 min. Digested peptides were desalted using SepPak C18 columns (Waters) according to manufacturer’s instructions.
Samples were analyzed by LC-MS/MS experiments on an LTQ Velos Pro Orbitrap mass spectrometer (Thermo, San Jose, CA) using the top twenty CID (collision-induced dissociation) method (Olsen et al., 2009). MS/MS spectra were searched against a composite database of the GFP:FUS-LC sequence and its reversed complement using the Sequest algorithm. Search parameters allowed for a dynamic modification of 79.966330 Da on Ser, Thr, and Tyr residues. Search results were filtered to include matches to the reverse data base by the linear discriminator function using parameters including Xcorr, dCN, missed cleavage, charge state (exclude 1+ peptides), mass accuracy, peptide length, and fraction of ions matched to MS/MS spectra (Huttlin et al., 2010). The identified phosphorylation sites were confirmed by targeted mass spectrometry identification. Localization of phosphorylation sites were assessed by the ModScore algorithm (Huttlin et al., 2010), in addition to manual evaluation of the quality of MS/MS spectra. To determine the extent of phosphorylation at the identified sites, the abundances of unphosphorylated peptides were quantified by integrating the peak areas in the corresponding extracted ion chromatograms for each time point. Then, the fractions of unphosphorylated peptides at the 30 or 60 min time points relative to the 0 min time point were calculated. The abundances of phosphorylated peptides were represented as 100% minus the fractions of corresponding unphosphorylated peptides.
For experiments in Fig. 5B, His-tagged FUS-LC was phosphorylated by DNA-PK in a 50 µl reaction volume containing 50 mM HEPES, pH 7.5, 100 mM KCl, 10 mM MgCl2, 0.2 mM EGTA, 0.1 mM EDTA, 1 mM DTT, 0.1 mM PMSF, 200 µM cold ATP, 1.2 µCi [γ-32P] ATP, 20 µM FUS-LC, 10 µg/ml calf thymus DNA, and 100 units of DNA-PK. Reactions proceeded for designated times at 30° C and were stopped by addition of SDS-PAGE running buffer, followed by SDS-PAGE and autoradiography. Band intensities were quantified with ImageJ software (https://imagej.nih.gov/ij).
Effects ofphosphorylation on hydrogel binding
Hydrogel droplets of mCherry:FUS-LC hydrogels were prepared as described previously (Kato et al., 2012). Phosphorylation of GFP:FUS-LC was performed as described previously (Han et al., 2012), with the following modifications. Reaction mixtures (60 µl) contained 50 mM HEPES, pH 7.5, 100 mM KCl, 1 mM DTT, 0.1 mM EDTA, 0.2 mM EGTA, 1 mM MgCl2, 1.5 µg calf thymus DNA, 400 µM ATP, 5 µM GFP:FUS-LC, and 600 units of DNA-PK (Promega, USA). Control reactions contained the same components, but without DNA-PK. These mixtures were incubated at 30° C for 60 min, then diluted with 600 µl of gelation buffer containing 20 mM Tris-HCl, pH 7.5, 200 mM NaCl, 20 mM BME, 0.5 mM EDTA, and 0.1 mM PMSF, then poured into dishes containing mCherry:FUS-LC hydrogels. The dishes were sealed with parafilm and stored at 4° C for 24 hours. Hydrogels were scanned by a confocal fluorescent microscope (Leica SP5) for GFP signals from bound proteins. Intensities of GFP fluorescence signals were analyzed with ImageJ as described in detail elsewhere (Xiang et al., 2015; Kato et al., 2017)
Effects of phosphorylation on droplet formation
A 96-well plate (USA Scientific, Orlando FL) was coated by 5% BSA for 2 h and dried overnight. His6-K-FUS-LC was purified through a Ni-NTA column, using buffer with 6 M Gdn-HCl as described above. After purification, the protein buffer was exchanged to 25 mM HEPES, pH 7.5, 5 mM BME, 2 M urea, using centrifugal filters (Millipore, Darmstadt, Germany). The protein concentration was adjusted to 55 mg/ml. FUS-LC liquid-like droplets were formed at room temperature by a rapid 30-fold dilution into DNA-PK reaction mixture (50 mM HEPES, pH 7.5, 100 mM KCl, 10 mM MgCl2, 0.2 mM EGTA, 0.1 mM EDTA, 1 mM DTT, 0.2 mM ATP, 10 µg/ml calf thymus DNA, 100 units of DNA-PK). Control experiments used the same mixture, but without either DNA-PK or ATP. Liquid-like droplets formed immediately in both the presence and the absence of DNA-PK. Optical images of droplets were taken with a Bio-Rad ZOE fluorescent cell imager (Bio-Rad, Hercules, CA), starting 0.5 h after dilution of FUS-LC into the reaction mixture and at successive 0.5 h intervals up to 2.0 h. The total area of liquid droplets in the images was measured with ImageJ.
QUANTIFICATION AND STATISTICAL ANALYSIS
Hydrogel binding experiments in Fig. 6B were repeated 2–3 times for each sample. Liquid-like droplet melting experiments in Fig. 6C were repeated twice for most double mutants and three times for the wild-type protein and the S42/S84 and S30/S61 mutants. Consistent results were obtained in repeated experiments. Error bars in Fig. 6E are standard deviations from hydrogel binding measurements on 3–4 hydrogels in a single experiment for each condition and standard deviations from measurements of droplet areas in three image sections for each condition. The correlation coefficient (R) from the plot in Fig. 6E was calculated using the GraphPad Prism 6 program.
DATA AND SOFTWARE AVAILABILITY
Atomic coordinates for 20 structural models for the FUS-LC fibril core have been deposited in the Protein Data Bank with accession code 5W3N. Solid state NMR chemical shifts have been deposited in the Biological Magnetic Resonance Data Bank with accession code 30304. 2D and 3D solid state NMR spectra of FUS-LC fibrils in Sparky format are available at http://dx.doi.org/10.17632/rxbh442x2g.1. Source code and instructions for the mcassign2b and mcassign2c programs are available upon request by e-mail to robertty@mail.nih.gov. Mass spectrometry data to identify phosphorylation sites of FUS-LC are available upon request by e-mail to steven.mcknight@utsouthwestern.edu.
Supplementary Material
(A) Thioflavin T fluorescence emission spectra of soluble FUS-LC (blue line) and FUS-LC fibrils (red line), showing enhanced fluorescence at 500 nm when ThT binds to FUS-LC fibrils. The peak at 423 nm arises from scattering of the excitation light. Emission spectra before addition of soluble FUS-LC (cyan line) or FUS-LC fibrils (magenta line) to the ThT solution are also shown. (B) Visible absorption spectra of Congo Red solutions containing soluble FUS-LC (blue line), FUS-LC fibrils (red line), or no FUS-LC (green line). Binding of Congo Red to FUS-LC fibrils leads to enhanced absorbance and a shift toward longer wavelengths. (C) Additional dark-field TEM image used for MPL measurement. Yellow and green rectangles represent typical regions for integration of image intensities from fibril or TMV segments (solid lines) and from background (dashed lines). (D) Histogram of MPL errors due to random variations in background intensities in dark-field TEM images. Values are calculated as described in Supplemental Methods. The best-fit Gaussian function, shown in red, has an 12 kDa/nm full-width-at-half-maximum, consistent with the width of the MPL peak in Fig. 1E.
Molecular dynamics simulation of a full-length FUS-LC fibril, including the structured core and dynamically disordered N- and C-terminal segments, Related to Fig. 4.
Molecular dynamics simulation of the FUS-LC fibril core in water, demonstrating stability of a pentameric assembly, Related to Fig. 4.
Representative 2D 13C-13C planes from 3D NCACX (red), NCOCX (blue), CONCA (green), and CANCX (purple) spectra, at the indicated 15N chemical shifts, are shown with crosspeak assignments from MCASSIGN calculations. 3D NCACX and NCOCX spectra were obtained at 21.1 T with MAS at 13.8 kHz. The 3D CONCA and CANCX spectra were obtained at 14.1 T with MAS at 12.0 kHz. Contour levels increase by successive factors of 1.3. (Note that crosspeaks within approximately ±0.6 ppm of the indicated 15N chemical shifts can appear in these planes, due to the non-zero 15N linewidths.)
(A) Number of times that signals from 3D NCACX (red circles), NCOCX (blue squares), and CONCA (green triangles) solid state NMR spectra of U-FUS-LC fibrils were assigned to residues 1–100 of the FUS-LC sequence in 50 independent MCASSIGN calculations (final round from the first set of calculations, as described in Methods Details). Filled symbols indicate that the assigned signals were identical in all calculations. Open symbols indicate that different signals were assigned in different calculations. Definite assignments for residues 53 and 54 were obtained in the second set of MCASSIGN calculations. (B) Signal-to-noise ratios for assigned crosspeaks from the 3D CONCA spectrum of U-FUS-LC fibrils (as reported by Sparky software), illustrating the large variations in crosspeak amplitudes attributable to variations in amplitudes and/or time scales of molecular motion within the FUS-LC fibril core. (C) Signal-to-noise ratios for unassigned crosspeaks from the 3D CONCA spectrum of U-FUS-LC fibrils. (D) Strip plots from 3D NCACX NCOCX, and CONCA spectra (red, blue, and green contours, respectively), showing the connections among crosspeaks in these spectra for residues 77–81).
(A) Schematic of the ligation reaction to produce segmentally labeled FUS-LC. A fusion protein comprised of the His-tagged N-terminal segment of FUS-LC, with a Cys substitution at residue X, and the AvaDnaE intein reacts with sodium 2-mercaptoethanesulfonate (MES) to produce the His-tagged N-terminal segment of FUS-LC with a thioester linkage to MES at its C-terminus. A fusion protein comprised of His-tagged mCherry and the C-terminal segment of FUS-LC, with a Cys substitution at residue X and an intervening Caspase-3 cleavage site sequence, is cleaved by Caspase-3 to produce the C-terminal segment of FUS-LC with a Cys substitution at its N-terminus. The two segments of FUS-LC react to produce full-length His-tagged FUS-LC, with a single Cys substitution at residue X. (B) SDS-PAGE analysis of FUS-LC constructs generated by ligation reactions with boundaries at residues 60 and 112. Full-length wild-type FUS-LC is shown as a reference (arrow). (C) TEM images of negatively-stained FUS-LC fibrils with segmental isotopic labeling. Variations in apparent fibril morphologies are attributable to differences in fibril growth conditions.
(A) Schematic representation of the N60-FUS-LC and C60-FUS-LC, with a Gln-to-Cys substitution at residue 60. (B) 1D 13C NMR spectra of N60-FUS-LC fibrils. Spectra were obtained with either 1H-13C cross-polarization (top) or 1H-13C INEPT (bottom) and are plotted with the same vertical scale, after correcting for differences in the number of scans. (C) Same as panel B, but for C60-FUS-LC fibrils. Spectra obtained with cross-polarization (CP) show signals from immobilized, structurally ordered sites, while spectra obtained with INEPT show signals from highly flexible, dynamic sites. (D,E) 2D 13C-13C NMR spectra of N60- and C60-FUS-LC fibrils, recorded at 14.1 T with MAS at 13.6 kHz, high-power 1H decoupling, and 50 ms DARR mixing periods between t1 and t2 dimensions. (F,G) 2D 15N-13C NMR spectra of N60- and C60-FUS-LC fibrils, recorded at 17.5 T with MAS at 17.0 kHz, high-power 1H decoupling, and 4 ms 15N-13Cα cross-polarization periods between t1 and t2 dimensions. Contour level increment factors are 1.3 in panels D-F and 1.25 in panel G. Black crosses in panels D-G indicate expected crosspeak positions from residues with definite chemical shift assignments (Table S1). Note that signals from some of these residues are too weak to be detected in these 2D spectra. Cyan crosses indicate expected crosspeak positions from signals that were observed in spectra of U-FUS-LC fibrils, but could not be assigned definitely to specific residues (Table S2).
(A) Backbone ϕ and ψ torsion angle predictions from TALOS-N for the core of FUS-LC fibrils, based on assigned solid state NMR chemical shifts. Error bars are uncertainties reported by TALOS-N. Closed and open symbols indicate predictions classified by TALOS-N as "strong" and "generous", respectively. (B) Intermolecular 13C-13C dipole-dipole couplings in FUS-LC fibrils prepared with 13C-labeled backbone carbonyl sites of all Thr or all Tyr residues. Open circles are data from PITHIRDS-CT measurements, with error bars representing uncertainty due to the root-mean-squared (rms) noise in the experimental spectra. Lines are simulations for linear chains of 13C nuclei with the indicated spacings. (C) Intramolecular 15N-15N dipole-dipole couplings from 15N-BARE measurements on 2-Glyc-FUS-LC fibrils. Examples of the 15N-BARE data are shown, with error bars representing uncertainty due to the rms noise in 2D spectra from which these data were obtained. Color-coded lines are simulations based on coordinates in the lowest-energy structural model for the FUS-LC fibril core. (D) Aliphatic region of a 2D 13C-13C DARR spectrum of 2-Glyc-FUS-LC fibrils, obtained with a 300 ms DARR mixing period. Long-range and short-range inter-residue crosspeaks are labeled in black and purple, respectively. (E) Sections of 2D planes from a 3D NCACX spectrum of 2-Glyc-FUS-LC fibrils (red contours, 400 ms DARR mixing period) and a 3D NCOCX spectrum of 1,3-Glyc-FUS-LC fibrils (blue contours, 400 ms DARR mixing period). (F) Distance restraints from long-range inter-residue crosspeaks, superimposed on the final structural model. Solid red lines are restraints with unambiguous assignments before structure calculations. Dashed red lines are a subset of the restraints with initially ambiguous assignments.
Predictions of amyloid-forming segments were obtained with the Aggrescan, FISH, FoldAmyloid, ZipperDB, MetAmyl, PASTA 2.0, WALTZ, and Zyggregator algorithms. Dashed blue lines indicate lower-bound thresholds for positive amyloid predictions. For ZipperDB, the dashed cyan line indicates the upper-bound threshold and dotted red lines span segments where proline residues preclude a prediction.
Highlights.
-
-
A specific 57-residue segment forms the core of FUS low complexity (FUS-LC) fibrils
-
-
Solid state NMR and segmental isotopic labeling define the FUS-LC core structure
-
-
Phosphorylation disrupts hydrogel binding and liquid droplet formation by FUS-LC
-
-
Phosphorylation sites map to the region of FUS-LC that forms the fibril core
Acknowledgments
We thank Drs. Matthew Pratt at University of Southern California and Tom Muir at Princeton University for providing AvaDnaE intein constructs. This work was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health. D.T.M. was supported by a Postdoctoral Research Associate (PRAT) fellowship from the National Institute of General Medical Sciences (NIGMS), award number 1Fi2GM117604-01. Structure and assignment calculations and molecular dynamics simulations used the high-performance computing resources of the NIH Biowulf cluster. Solid state NMR data at 21.1 T were acquired under project P08483 through the Users Program of NHMFL, which is supported by NSF DMR-1157490 and the State of Florida. We thank Dr. Charles D. Schwieters for assistance with structure calculations. Work performed at UTSWMC was supported by grant 5U01GM107623-03 from the NIGMS and unrestricted funds provided to S.L.M. by an anonymous donor. We thank Yonghao Yu and Leeju Wu for technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
D.T.M, M.K., Y.L., S.L.M., and R.T. planned experiments. D.T.M., M.K., Y.L., K.R.T., I.H., and R.T. conducted experiments and analyzed data. D.T.M., M.K., Y.L., S.L.M., and R.T. wrote the manuscript.
References
- Alberti S, Hyman AA. Are aberrant phase transitions a driver of cellular aging? Bioessays. 2016;38:959–968. doi: 10.1002/bies.201600042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani SF, Lee HO, Hyman AA, Rosen MK. Biomolecular condensates: Organizers of cellular biochemistry. Nat Rev Mol Cell Biol. 2017;18:285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayro MJ, Chen B, Yau WM, Tycko R. Site-specific structural variations accompanying tubular assembly of the HIV-1 capsid protein. J Mol Biol. 2014;426:1109–1127. doi: 10.1016/j.jmb.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett AE, Rienstra CM, Auger M, Lakshmi KV, Griffin RG. Heteronuclear decoupling in rotating solids. J Chem Phys. 1995;103:6951–6958. [Google Scholar]
- Bertini I, Gonnelli L, Luchinat C, Mao JF, Nesi A. A new structural model of Aβ(40) fibrils. J Am Chem Soc. 2011;133:16013–16022. doi: 10.1021/ja2035859. [DOI] [PubMed] [Google Scholar]
- Burke KA, Janke AM, Rhine CL, Fawzi NL. Residue-by-residue view of in vitro FUS granules that bind the C-terminal domain of RNA polymerase II. Mol Cell. 2015;60:231–241. doi: 10.1016/j.molcel.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellani F, van Rossum BJ, Diehl A, Rehbein K, Oschkinat H. Determination of solid state NMR structures of proteins by means of three-dimensional 15N-13C-13C dipolar correlation spectroscopy and chemical shift analysis. Biochemistry. 2003;42:11476–11483. doi: 10.1021/bi034903r. [DOI] [PubMed] [Google Scholar]
- Chen B, Thurber KR, Shewmaker F, Wickner RB, Tycko R. Measurement of amyloid fibril mass-per-length by tilted-beam transmission electron microscopy. Proc Natl Acad Sci U S A. 2009;106:14339–14344. doi: 10.1073/pnas.0907821106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin MT, Silvers R, Ni QZ, Can TV, Sergeyev I, Rosay M, Donovan KJ, Michael B, Wall J, Linse S, et al. Atomic resolution structure of monomorphic Aβ(42) amyloid fibrils. J Am Chem Soc. 2016;138:9663–9674. doi: 10.1021/jacs.6b05129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot NS, Castillo V, Grana-Montes R, Zamora SV. Aggrescan: Method, application, and perspectives for drug design. In: Baron R, editor. Computational drug discovery and design. 2012. pp. 199–220. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRpipe: A multidimensional spectral processing system based on Unix pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Deng QD, Holler CJ, Taylor G, Hudson KF, Watkins W, Gearing M, Ito D, Murray ME, Dickson DW, Seyfned NT, et al. FUS is phosphorylated by DNA-PK and accumulates in the cytoplasm after DNA damage. J Neurosci. 2014;34:7802–7813. doi: 10.1523/JNEUROSCI.0172-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- Elbaum-Garfinkle S, Kim Y, Szczepaniak K, Chen CCH, Eckmann CR, Myong S, Brangwynne CP. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc Natl Acad Sci USA. 2015;112:7189–7194. doi: 10.1073/pnas.1504822112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emily M, Talvas A, Delamarche C. Metamyl: A meta-predictor for amyloid proteins. PLoS One. 2013:8. doi: 10.1371/journal.pone.0079722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, Richardson TM, Kriwacki RW, Pappu RV, Brangwynne CP. Coexisting liquid phases underlie nucleolar subcompartments. Cell. 2016;165:1686–1697. doi: 10.1016/j.cell.2016.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbuzynskiy SO, Lobanov MY, Galzitskaya OV. FoldAmyloid: A method of prediction of amyloidogenic regions from protein sequence. Bioinformatics. 2010;26:326–332. doi: 10.1093/bioinformatics/btp691. [DOI] [PubMed] [Google Scholar]
- Gardiner M, Toth R, Vandermoere F, Morrice NA, Rouse J. Identification and characterization of FUS/TLS as a new target of ATM. Biochem J. 2008;415:297–307. doi: 10.1042/BJ20081135. [DOI] [PubMed] [Google Scholar]
- Gasior P, Kotulska M. Fish amyloid: A new method for finding amyloidogenic segments in proteins based on site specific co-occurence of aminoacids. BMC Bioinformatics. 2014:15. doi: 10.1186/1471-2105-15-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han TNW, Kato M, Xie SH, Wu LC, Mirzaei H, Pei JM, Chen M, Xie Y, Allen I, Xiao GH, et al. Cell-free formation of RNA granules: Bound RNAs identify features and components of cellular assemblies. Cell. 2012;149:768–779. doi: 10.1016/j.cell.2012.04.016. [DOI] [PubMed] [Google Scholar]
- Harrison AF, Shorter J. RNA-binding proteins with prion-like domains in health and disease. Biochem J. 2017;474:1417–1438. doi: 10.1042/BCJ20160499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heise H, Hoyer W, Becker S, Andronesi OC, Riedel D, Baldus M. Molecular-level secondary structure, polymorphism, and dynamics of full-length α-synuclein fibrils studied by solid state NMR. Proc Natl Acad Sci U S A. 2005;102:15871–15876. doi: 10.1073/pnas.0506109102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong M, Jakes K. Selective and extensive 13C labeling of a membrane protein for solid state NMR investigations. J Biomol NMR. 1999;14:71–74. doi: 10.1023/a:1008334930603. [DOI] [PubMed] [Google Scholar]
- Hu KN, Qiang W, Bermejo GA, Schwieters CD, Tycko R. Restraints on backbone conformations in solid state NMR studies of uniformly labeled proteins from quantitative amide 15N-15N and carbonyl 13C-13C dipolar recoupling data. J Magn Reson. 2012;218:115–127. doi: 10.1016/j.jmr.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu KN, Qiang W, Tycko R. A general Monte carlo/simulated annealing algorithm for resonance assignment in NMR of uniformly labeled biopolymers. J Biomol NMR. 2011;50:267–276. doi: 10.1007/s10858-011-9517-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villen J, Haas W, Sowa ME, Gygi SP. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Han TNW, Xie SH, Shi K, Du XL, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei JM, et al. Cell-free formation of RNA granules: Low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiebler MA, Bassell GJ. Neuronal RNA granules: Movers and makers. Neuron. 2006;51:685–690. doi: 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- Kwon I, Kato M, Xiang SH, Wu L, Theodoropoulos P, Mirzaei H, Han T, Xie SH, Corden JL, McKnight SL. Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell. 2013;155:1049–1060. doi: 10.1016/j.cell.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond AI, Spector DL. Nuclear speckles: A model for nuclear organelles. Nat Rev Mol Cell Biol. 2003;4:605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- Lin Y, Mori E, Kato M, Xiang SH, Wu LJ, Kwon I, McKmght SL. Toxic PR poly-dipeptides encoded by the C9orf72 repeat expansion target LC domain polymers. Cell. 2016;167:789−+. doi: 10.1016/j.cell.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Protter DSW, Rosen MK, Parker R. Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol Cell. 2015;60:208–219. doi: 10.1016/j.molcel.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JG, Perumal NB, Oldfield CJ, Su EW, Uversky VN, Dunker AK. Intrinsic disorder in transcription factors. Biochemistry. 2006;45:6873–6888. doi: 10.1021/bi0602718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu JX, Qiang W, Yau WM, Schwieters CD, Meredith SC, Tycko R. Molecular structure of β-amyloid fibrils in Alzheimer's disease brain tissue. Cell. 2013;154:1257–1268. doi: 10.1016/j.cell.2013.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer-Stroh S, Debulpaep M, Kuemmerer N, de la Paz ML, Martins IC, Reumers J, Morris KL, Copland A, Serpell L, Serrano L, et al. Exploring the sequence determinants of amyloid structure using position-specific scoring matrices. Nat Methods. 2010;7:237–U109. doi: 10.1038/nmeth.1432. [DOI] [PubMed] [Google Scholar]
- Morris GA, Freeman R. Enhancement of nuclear magnetic resonance signals by polarization transfer. J Am Chem Soc. 1979;101:760–762. [Google Scholar]
- Murakami T, Qamar S, Lin JQ, Schierle GSK, Rees E, Miyashita A, Costa AR, Dodd RB, Chan FTS, Michel CH, et al. ALS/FTD mutation-induced phase transition of FUS liquid droplets and reversible hydrogels into irreversible hydrogels impairs RNP granule function. Neuron. 2015;88:678–690. doi: 10.1016/j.neuron.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott TJ, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A, Craggs TD, Bazett-Jones DP, Pawson T, Forman-Kay JD, et al. Phase transition of a disordered uage protein generates environmentally responsive membraneless organelles. Mol Cell. 2015;57:936–947. doi: 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JV, Schwartz JC, Griep-Raming J, Nielsen ML, Damoc E, Denisov E, Lange O, Remes P, Taylor D, Splendore M, et al. A dual pressure linear ion trap orbitrap instrument with very high sequencing speed. Mol Cell Proteomics. 2009;8:2759–2769. doi: 10.1074/mcp.M900375-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paravastu AK, Leapman RD, Yau WM, Tycko R. Molecular structural basis for polymorphism in Alzheimer's β-amyloid fibrils. Proc Natl Acad Sci USA. 2008;105:18349–18354. doi: 10.1073/pnas.0806270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, et al. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- Perutz MF, Johnson T, Suzuki M, Finch JT. Glutamine repeats as polar zippers: Their possible role in inherited neurodegenerative diseases. Proc Natl Acad Sci U S A. 1994;91:5355–5358. doi: 10.1073/pnas.91.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines A, Gibby MG, Waugh JS. Proton-enhanced NMR of dilute spins in solids. J Chem Phys. 1973;59:569–590. [Google Scholar]
- Saha S, Weber CA, Nousch M, Adame-Arana O, Hoege C, Hein MY, Osborne-Nishimura E, Mahamid J, Jahnel M, Jawerth L, et al. Polar positioning of phase-separated liquid compartments in cells regulated by an mRNA competition mechanism. Cell. 2016;166:1572–1584. doi: 10.1016/j.cell.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJW, McFarlane HT, et al. Atomic structures of amyloid cross-β spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- Schutz AK, Vagt T, Huber M, Ovchinnikova OY, Cadalbert R, Wall J, Guntert P, Bockmann A, Glockshuber R, Meier BH. Atomic-resolution three-dimensional structure of amyloid-β fibrils bearing the Osaka mutation. Angew Chem-Int Edit. 2015;54:331–335. doi: 10.1002/anie.201408598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwieters CD, Kuszewski JJ, Clore GM. Using Xplor-NIH for NMR molecular structure determination. Prog Nucl Magn Reson Spectrosc. 2006;48:47–62. [Google Scholar]
- Shah NH, Dann GP, Vila-Perello M, Liu ZH, Muir TW. Ultrafast protein splicing is common among cyanobacterial split inteins: Implications for protein enGlneering. J Am Chem Soc. 2012;734:11338–11341. doi: 10.1021/ja303226x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield P, Garrard S, Derewenda Z. Overcoming expression and purification problems of rhogdi using a family of "parallel" expression vectors. Protein Expr Purif. 1999;15:34–39. doi: 10.1006/prep.1998.1003. [DOI] [PubMed] [Google Scholar]
- Shen Y, Bax A. Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J Biomol NMR. 2013;56:227–241. doi: 10.1007/s10858-013-9741-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi KY, Mori E, Nizami ZF, Lin Y, Kato M, Xiang SH, Wu LC, Ding M, Yu YH, Gall JG, et al. Toxic PRn poly-dipeptides encoded by the C9orf72 repeat expansion block nuclear import and export. Proc Natl Acad Sci U S A. 2017;114:E1111–E1117. doi: 10.1073/pnas.1620293114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strambio-De-Castillia C, Niepel M, Rout MP. The nuclear pore complex: BridGlng nuclear transport and gene regulation. Nat Rev Mol Cell Biol. 2010;11:490–501. doi: 10.1038/nrm2928. [DOI] [PubMed] [Google Scholar]
- Takegoshi K, Nakamura S, Terao T. 13C-1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem Phys Lett. 2001;344:631–637. [Google Scholar]
- Tartaglia GG, Vendruscolo M. The Zyggregator method for predicting protein aggregation propensities. Chem Soc Rev. 2008;37:1395–1401. doi: 10.1039/b706784b. [DOI] [PubMed] [Google Scholar]
- Taylor JP, Brown RH, Cleveland DW. Decoding ALS: From genes to mechanism. Nature. 2016;539:197–206. doi: 10.1038/nature20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirumalai D, Reddy G, Straub JE. Role of water in protein aggregation and amyloid polymorphism. Accounts Chem Res. 2012;45:83–92. doi: 10.1021/ar2000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MJ, Sievers SA, Karanicolas J, Ivanova MI, Baker D, Eisenberg D. The 3D profile method for identifying fibril-forming segments of proteins. Proc Natl Acad Sci U S A. 2006;103:4074–4078. doi: 10.1073/pnas.0511295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuttle MD, Comellas G, Nieuwkoop AJ, Covell DJ, Berthold DA, Kloepper KD, Courtney JM, Kim JK, Barclay AM, Kendall A, et al. Solid state NMR structure of a pathogenic fibril of full-length human α-synuclein. Nat Struct Mol Biol. 2016;23:409–415. doi: 10.1038/nsmb.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycko R. Symmetry-based constant-time homonuclear dipolar recoupling in solid state NMR. J Chem Phys. 2007:126. doi: 10.1063/1.2437194. [DOI] [PubMed] [Google Scholar]
- Van Melckebeke H, Wasmer C, Lange A, Ab E, Loquet A, Bockmann A, Meier BH. Atomic-resolution three-dimensional structure of HET-s(218–289) amyloid fibrils by solid state NMR spectroscopy. J Am Chem Soc. 2010;132:13765–13775. doi: 10.1021/ja104213j. [DOI] [PubMed] [Google Scholar]
- Walsh I, Seno F, Tosatto SCE, Trovato A. PASTA 2.0: An improved server for protein aggregation prediction. Nucleic Acids Res. 2014;42:W301–W307. doi: 10.1093/nar/gku399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walti MA, Ravotti F, Arai H, Glabe CG, Wall JS, Bockmann A, Guntert P, Meier BH, Riek R. Atomic-resolution structure of a disease-relevant Aβ(1–42) amyloid fibril. Proc Natl Acad Sci U S A. 2016;113:E4976–E4984. doi: 10.1073/pnas.1600749113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang SH, Kato M, Wu LC, Lin Y, Ding M, Zhang YJ, Yu YH, McKnight SL. The LC domain of hnRNPA2 adopts similar conformations in hydrogel polymers, liquid-like droplets, and nuclei. Cell. 2015;163:829–839. doi: 10.1016/j.cell.2015.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao YL, Ma BY, McElheny D, Parthasarathy S, Long F, Hoshi M, Nussinov R, Ishii Y. Aβ(1-42) fibril structure illuminates self-recognition and replication of amyloid in Alzheimer's disease. Nat Struct Mol Biol. 2015;22:499–U497. doi: 10.1038/nsmb.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou HX. Loops, linkages, rings, catenanes, cages, and crowders: Entropy-based strategies for stabilizing proteins. Accounts Chem Res. 2004;37:123–130. doi: 10.1021/ar0302282. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Thioflavin T fluorescence emission spectra of soluble FUS-LC (blue line) and FUS-LC fibrils (red line), showing enhanced fluorescence at 500 nm when ThT binds to FUS-LC fibrils. The peak at 423 nm arises from scattering of the excitation light. Emission spectra before addition of soluble FUS-LC (cyan line) or FUS-LC fibrils (magenta line) to the ThT solution are also shown. (B) Visible absorption spectra of Congo Red solutions containing soluble FUS-LC (blue line), FUS-LC fibrils (red line), or no FUS-LC (green line). Binding of Congo Red to FUS-LC fibrils leads to enhanced absorbance and a shift toward longer wavelengths. (C) Additional dark-field TEM image used for MPL measurement. Yellow and green rectangles represent typical regions for integration of image intensities from fibril or TMV segments (solid lines) and from background (dashed lines). (D) Histogram of MPL errors due to random variations in background intensities in dark-field TEM images. Values are calculated as described in Supplemental Methods. The best-fit Gaussian function, shown in red, has an 12 kDa/nm full-width-at-half-maximum, consistent with the width of the MPL peak in Fig. 1E.
Molecular dynamics simulation of a full-length FUS-LC fibril, including the structured core and dynamically disordered N- and C-terminal segments, Related to Fig. 4.
Molecular dynamics simulation of the FUS-LC fibril core in water, demonstrating stability of a pentameric assembly, Related to Fig. 4.
Representative 2D 13C-13C planes from 3D NCACX (red), NCOCX (blue), CONCA (green), and CANCX (purple) spectra, at the indicated 15N chemical shifts, are shown with crosspeak assignments from MCASSIGN calculations. 3D NCACX and NCOCX spectra were obtained at 21.1 T with MAS at 13.8 kHz. The 3D CONCA and CANCX spectra were obtained at 14.1 T with MAS at 12.0 kHz. Contour levels increase by successive factors of 1.3. (Note that crosspeaks within approximately ±0.6 ppm of the indicated 15N chemical shifts can appear in these planes, due to the non-zero 15N linewidths.)
(A) Number of times that signals from 3D NCACX (red circles), NCOCX (blue squares), and CONCA (green triangles) solid state NMR spectra of U-FUS-LC fibrils were assigned to residues 1–100 of the FUS-LC sequence in 50 independent MCASSIGN calculations (final round from the first set of calculations, as described in Methods Details). Filled symbols indicate that the assigned signals were identical in all calculations. Open symbols indicate that different signals were assigned in different calculations. Definite assignments for residues 53 and 54 were obtained in the second set of MCASSIGN calculations. (B) Signal-to-noise ratios for assigned crosspeaks from the 3D CONCA spectrum of U-FUS-LC fibrils (as reported by Sparky software), illustrating the large variations in crosspeak amplitudes attributable to variations in amplitudes and/or time scales of molecular motion within the FUS-LC fibril core. (C) Signal-to-noise ratios for unassigned crosspeaks from the 3D CONCA spectrum of U-FUS-LC fibrils. (D) Strip plots from 3D NCACX NCOCX, and CONCA spectra (red, blue, and green contours, respectively), showing the connections among crosspeaks in these spectra for residues 77–81).
(A) Schematic of the ligation reaction to produce segmentally labeled FUS-LC. A fusion protein comprised of the His-tagged N-terminal segment of FUS-LC, with a Cys substitution at residue X, and the AvaDnaE intein reacts with sodium 2-mercaptoethanesulfonate (MES) to produce the His-tagged N-terminal segment of FUS-LC with a thioester linkage to MES at its C-terminus. A fusion protein comprised of His-tagged mCherry and the C-terminal segment of FUS-LC, with a Cys substitution at residue X and an intervening Caspase-3 cleavage site sequence, is cleaved by Caspase-3 to produce the C-terminal segment of FUS-LC with a Cys substitution at its N-terminus. The two segments of FUS-LC react to produce full-length His-tagged FUS-LC, with a single Cys substitution at residue X. (B) SDS-PAGE analysis of FUS-LC constructs generated by ligation reactions with boundaries at residues 60 and 112. Full-length wild-type FUS-LC is shown as a reference (arrow). (C) TEM images of negatively-stained FUS-LC fibrils with segmental isotopic labeling. Variations in apparent fibril morphologies are attributable to differences in fibril growth conditions.
(A) Schematic representation of the N60-FUS-LC and C60-FUS-LC, with a Gln-to-Cys substitution at residue 60. (B) 1D 13C NMR spectra of N60-FUS-LC fibrils. Spectra were obtained with either 1H-13C cross-polarization (top) or 1H-13C INEPT (bottom) and are plotted with the same vertical scale, after correcting for differences in the number of scans. (C) Same as panel B, but for C60-FUS-LC fibrils. Spectra obtained with cross-polarization (CP) show signals from immobilized, structurally ordered sites, while spectra obtained with INEPT show signals from highly flexible, dynamic sites. (D,E) 2D 13C-13C NMR spectra of N60- and C60-FUS-LC fibrils, recorded at 14.1 T with MAS at 13.6 kHz, high-power 1H decoupling, and 50 ms DARR mixing periods between t1 and t2 dimensions. (F,G) 2D 15N-13C NMR spectra of N60- and C60-FUS-LC fibrils, recorded at 17.5 T with MAS at 17.0 kHz, high-power 1H decoupling, and 4 ms 15N-13Cα cross-polarization periods between t1 and t2 dimensions. Contour level increment factors are 1.3 in panels D-F and 1.25 in panel G. Black crosses in panels D-G indicate expected crosspeak positions from residues with definite chemical shift assignments (Table S1). Note that signals from some of these residues are too weak to be detected in these 2D spectra. Cyan crosses indicate expected crosspeak positions from signals that were observed in spectra of U-FUS-LC fibrils, but could not be assigned definitely to specific residues (Table S2).
(A) Backbone ϕ and ψ torsion angle predictions from TALOS-N for the core of FUS-LC fibrils, based on assigned solid state NMR chemical shifts. Error bars are uncertainties reported by TALOS-N. Closed and open symbols indicate predictions classified by TALOS-N as "strong" and "generous", respectively. (B) Intermolecular 13C-13C dipole-dipole couplings in FUS-LC fibrils prepared with 13C-labeled backbone carbonyl sites of all Thr or all Tyr residues. Open circles are data from PITHIRDS-CT measurements, with error bars representing uncertainty due to the root-mean-squared (rms) noise in the experimental spectra. Lines are simulations for linear chains of 13C nuclei with the indicated spacings. (C) Intramolecular 15N-15N dipole-dipole couplings from 15N-BARE measurements on 2-Glyc-FUS-LC fibrils. Examples of the 15N-BARE data are shown, with error bars representing uncertainty due to the rms noise in 2D spectra from which these data were obtained. Color-coded lines are simulations based on coordinates in the lowest-energy structural model for the FUS-LC fibril core. (D) Aliphatic region of a 2D 13C-13C DARR spectrum of 2-Glyc-FUS-LC fibrils, obtained with a 300 ms DARR mixing period. Long-range and short-range inter-residue crosspeaks are labeled in black and purple, respectively. (E) Sections of 2D planes from a 3D NCACX spectrum of 2-Glyc-FUS-LC fibrils (red contours, 400 ms DARR mixing period) and a 3D NCOCX spectrum of 1,3-Glyc-FUS-LC fibrils (blue contours, 400 ms DARR mixing period). (F) Distance restraints from long-range inter-residue crosspeaks, superimposed on the final structural model. Solid red lines are restraints with unambiguous assignments before structure calculations. Dashed red lines are a subset of the restraints with initially ambiguous assignments.
Predictions of amyloid-forming segments were obtained with the Aggrescan, FISH, FoldAmyloid, ZipperDB, MetAmyl, PASTA 2.0, WALTZ, and Zyggregator algorithms. Dashed blue lines indicate lower-bound thresholds for positive amyloid predictions. For ZipperDB, the dashed cyan line indicates the upper-bound threshold and dotted red lines span segments where proline residues preclude a prediction.