Figure 1. Classes of photolyases and structures of CPD and 6-4 photolyases.
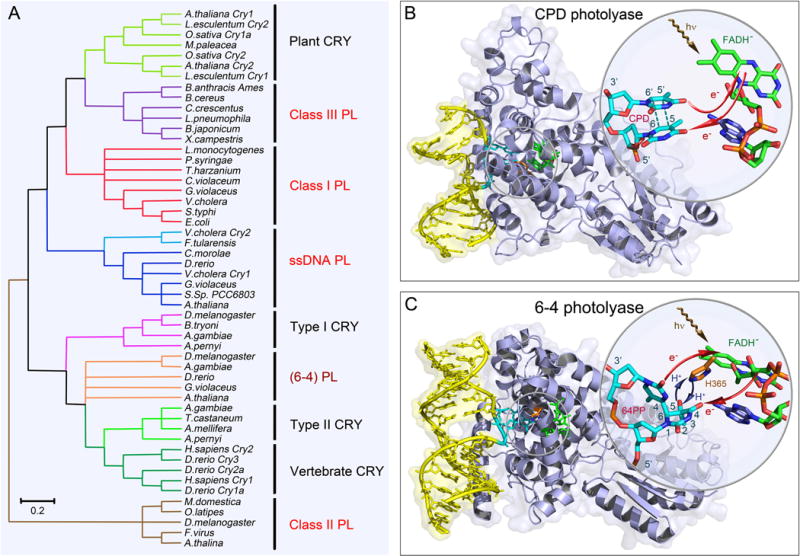
(A) The unrooted phylogenetic tree of the photolyase/cryptochrome protein family and representative members. Major classes in the family and their evolutionary relationships are shown. Photolyases (PLs) are divided into CPD PL and 6-4 PL based on their different DNA substrate. CPD PLs are further divided into class I, II, III and single-stranded DNA (ssDNA) PLs. The ssDNA PLs were previously classified as CRY with Cry-DASH designation. The class II PLs are distantly related to other classes. (B) X-ray complex structure of Anacystis nidulans CPD photolyase (PDB: 1TEZ) bound with double-stranded DNA (yellow) containing a thymine dimer after in situ repair. The thymine dimer is flipped out of DNA and inserts into the active site of protein. The close-up view shows the relative positions of the catalytic cofactor FADH− and the repaired substrate. The flavin cofactor is in the U-shaped configuration and the adenine moiety and the lumiflavin moiety have a short separation distance. The red arrows indicate the electron tunneling pathways in repair. (C) X-ray complex structure of Drosophila melanogaster (6-4) photolyase bound with double-stranded DNA (yellow) containing a (6-4) photoproduct (6-4PP). Like CPD, the 6-4PP is flipped out of DNA and inserts into the active site. The close-up view shows that the conserved histidine residue (orange) in the active site (H365 in D. melanogaster and H364 in Arabidopsis thaliana) is close to the 6-4PP. The blue arrow represents the proton transfer between the two during repair. Similar electron tunneling occurs in 6-4PLs as shown with the red arrows.
