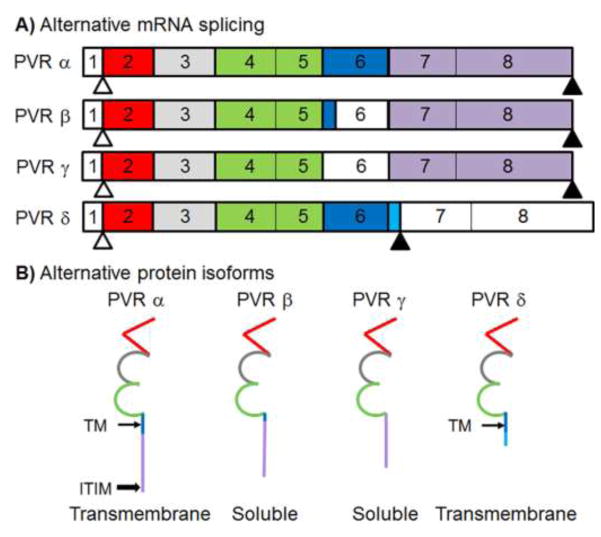
Fig 1. Schematic of exon map for PVR A) RNA spliceforms and B) protein isoforms.
Unshaded exons are not expressed in the protein. Exons are color coded to match the portions of the protein that they encode: red = Ig-like domain 1, grey = Ig-like domain 2, green = Ig-like domain 3, dark blue = transmembrane domain (thin arrow/TM), light blue = unique sequence in δ isoform, and gold = C-terminal domain. Start codons appear as white triangles and stop codons appear as shaded triangles. Compared to the canonical isoform PVRα, soluble isoforms PVRβ and γ contain splicing events in exon 6 which result in partial (β) or complete (γ) loss of exon 6. There is an alternative splicing event between transmembrane isoforms PVRα and PVRδ in which an additional eight residues and a stop codon are incorporated at the end of exon 6, resulting in exons 7 and 8 not being translated in PVRδ. The immunoreceptor tyrosine-based inhibitory motif (ITIM) of PVRα is indicated (block arrow).