Figure 4.
Crystal Structure of Ycg1–Brn1 in Complex with DNA
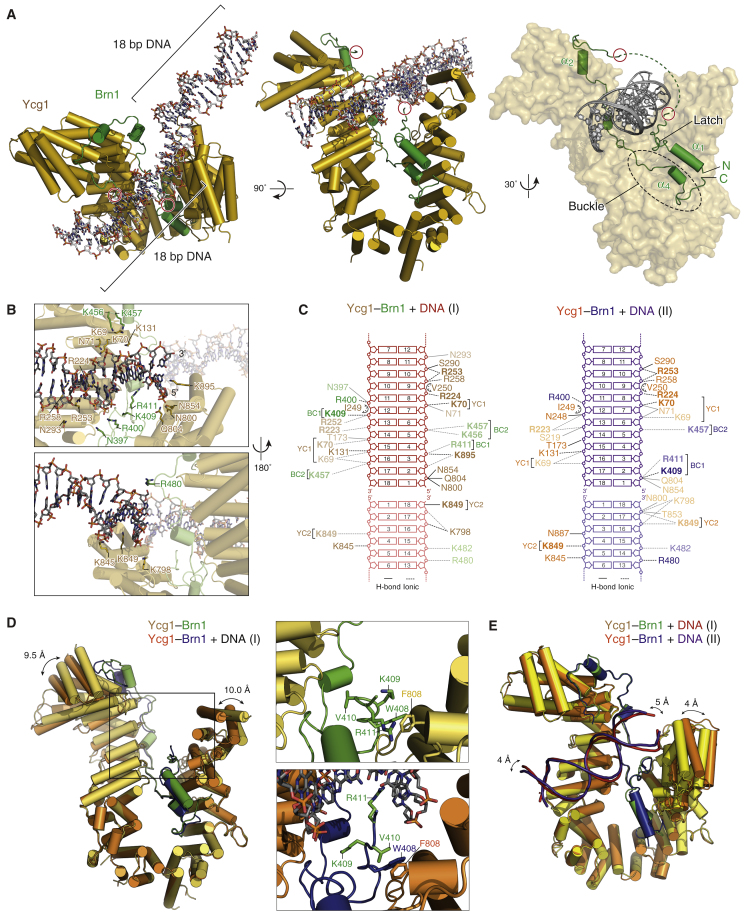
(A) Cartoon and surface models of the Sc Ycg1–Brn1 complex bound to an 18-bp dsDNA substrate and to the 18-bp dsDNA of its symmetry-related neighbor, based on the Sc Ycg16–498, 556–932–Brn1384–529 construct. The ends of the disordered region of Sc Brn1 (red circles), the Brn1 kleisin disordered linker, α4 buckle, and α1 latch regions are indicated.
(B) Close-up views of the Sc Ycg1–Brn1 interaction with DNA.
(C) Schematic illustrations of the main contacts of Sc Ycg1 and Brn1 residues with the 18-bp DNAs in crystal forms I and II. Highly conserved residues are shown in bold, distant residues are shown in opaque. The length of the lines is proportional to the observed distances of H-bonds (solid lines) or ionic interactions (dotted lines).
(D) Structural alignment of Sc Ycg1–Brn1 and Sc Ycg1–Brn1–DNA (crystal form I) using all Cα atoms (RMSD 1.95 Å over 820 Cα). Arrows indicate conformational differences in the N-terminal shoulder region of Ycg1. Close-up views highlight changes of side chains in the Brn1 latch region.
(E) Structural alignment of Sc Ycg1–Brn1–DNA crystal forms I and II DNA using all Cα atoms (RMSD 0.79 Å over 685 Cα). Arrows indicate conformational differences in the DNA and in the C-terminal shoulder region of Ycg1.
See also Figure S5.