Figure 5.
DNA Entrapment by the Kleisin Loop
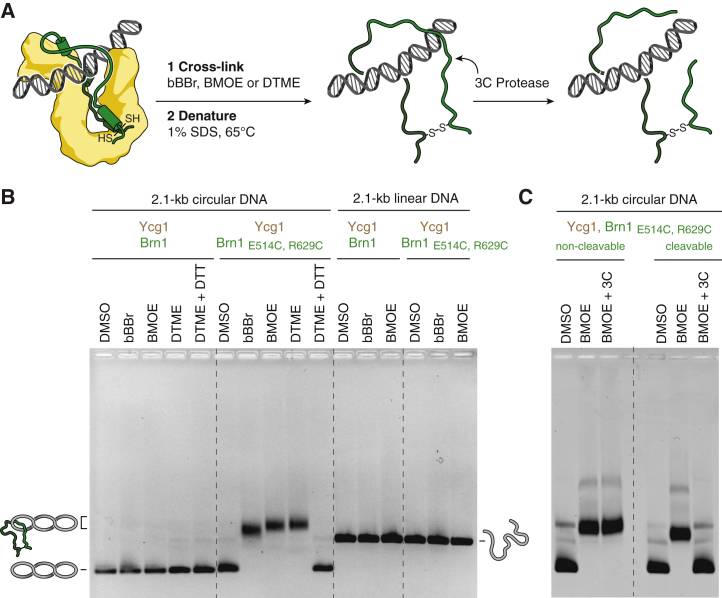
(A) Outline of the experimental setup to test whether the Brn1 loop encircles DNA.
(B) Copurified Ct Ycg124–1006–His6-TEV-Brn1515–634 subcomplexes without or with an additional cysteine pair engineered into the Brn1 latch and buckle regions (Ct Brn1E514C, R629C) were incubated with 2.1-kb circular or linear DNA substrates; DMSO solvent; or bBBr, BMOE, or DTME crosslinkers and denatured at 65°C in the presence of 1% SDS. Changes in DNA mobility were tested by EMSA and EtBr.
(C) EMSA of a 2.1-kb circular DNA using copurified Ct Ycg124–1006–His6-TEV-Brn1515–634 subcomplexes containing the Brn1E514C, R629C cysteine pair and a target site for HRV-3C protease engineered following residue P549 in the Ct Brn1 linker region (cleavable) or no-protease site (non-cleavable) as described in (B). Following addition of DNA and incubation with DMSO solvent or BMOE crosslinker, samples were treated with HRV-3C protease (±3C) or buffer only.
See also Figure S6.