Figure 6.
Kleisin Loop Closure Is Required for Condensin’s Loading onto Chromosomes and DNA-Dependent Stimulation of its ATPase
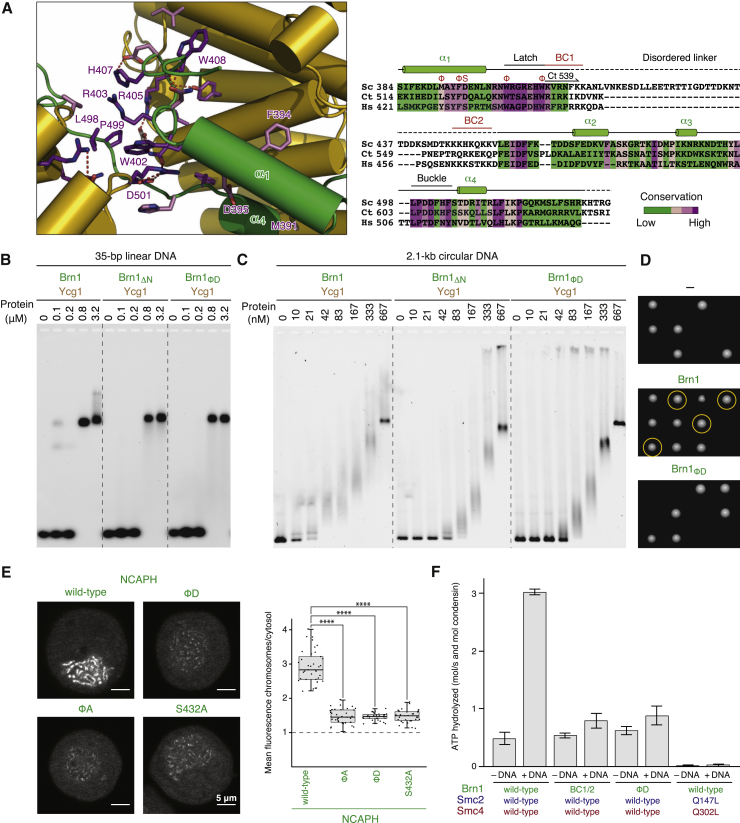
(A) Close-up view of the Sc Brn1 latch-buckle interaction. Conserved Brn1 residues are marked in purple, relevant H-bonds are indicated (dotted lines). Sequence alignment of the Ycg1-interacting region of Sc, Ct, and Hs γ-kleisins. Secondary structure elements are highlighted based on the Sc Ycg1–Brn1 structure. Conserved latch and buckle regions, DNA binding site mutations (BC1 and BC2), mutations of hydrophobic latch residues (Φ), and the phosphorylated serine residue in Hs NCAPHS432 are highlighted.
(B) EMSA of a 35-bp 6-FAM-labeled linear DNA (0.2 μM) with copurified wild-type (Ct Ycg124-1006–His6-TEV-Brn1515–634), Brn1ΔN truncated (Ct Ycg124–1006–His6-TEV-Brn1539–634), and Brn1ΦD latch mutant (Ct Ycg124–1006–His6-TEV-Brn1515–634, L521D, F524D, W532D, W538D) subcomplexes detected by EtBr staining.
(C) EMSA of a 2.1-kb circular DNA (10 nM) with the same proteins as described in (B) detected by EtBr staining. Note the differences in the fraction of non-shifted DNA.
(D) Tetrad dissection of BRN1/brn1Δ diploid budding yeast cells (strains C4237, C4239, C4895) expressing ectopic copies of Brn1 wild-type or Brn1ΦD (Brn1M391D, F394D, W402D, W408D) mutant versions of versions of Brn1-PK6. Images were recorded after three days at 25°C. Genetic marker analysis identifies BRN1x, brn1Δ cells (circles).
(E) Representative example images of nocodazole-arrested HeLa cells expressing mCherry-tagged histone H2B and transiently transfected EGFP-tagged NCAPH as wild-type, latch mutant (ΦA: Hs NCAPHY428A, F431A, W439A, W445A, ΦD: NCAPHY428D, F431D, W439D, W445D), or non-phosphorylatable latch (Hs NCAPHS432A) versions. Scale bars: 5 μm. The graph plots ratios of chromosomal to cytosolic EGFP intensities. Horizontal lines indicate median, hinges indicate first and third quartiles, and whiskers extend to the highest or lowest point from the hinges within 1.5 times inter-quartile range, calculated from two independent experiments with a total of n = 37 (NCAPH), n = 37 (NCAPHΦA), n = 31 (NCAPHΦD), and n = 31 (NCAPHS432A) cells (p < 0.0001 by Student’s t test with Welch’s correction).
(F) ATP hydrolysis by copurified Sc condensin holocomplexes (0.5 μM, Smc2–Smc4-StrepII3–Ycs4–Ycg1–Brn1-His12-HA3) containing wild-type, Brn1BC1/2 DNA binding (Sc Brn1K409D, R411D, K414D, K451D, K452D, K454D, K456D, K457D), Brn1ΦD latch (Sc Brn1M391D, F394D, W402D, W408D), or ATPase deficient (Sc Smc2Q147L–Smc4Q302L) mutant versions with and without a 6.4-kb relaxed circular DNA at saturated ATP concentrations (5 mM). The plot shows mean ± SD from three independent experiments.