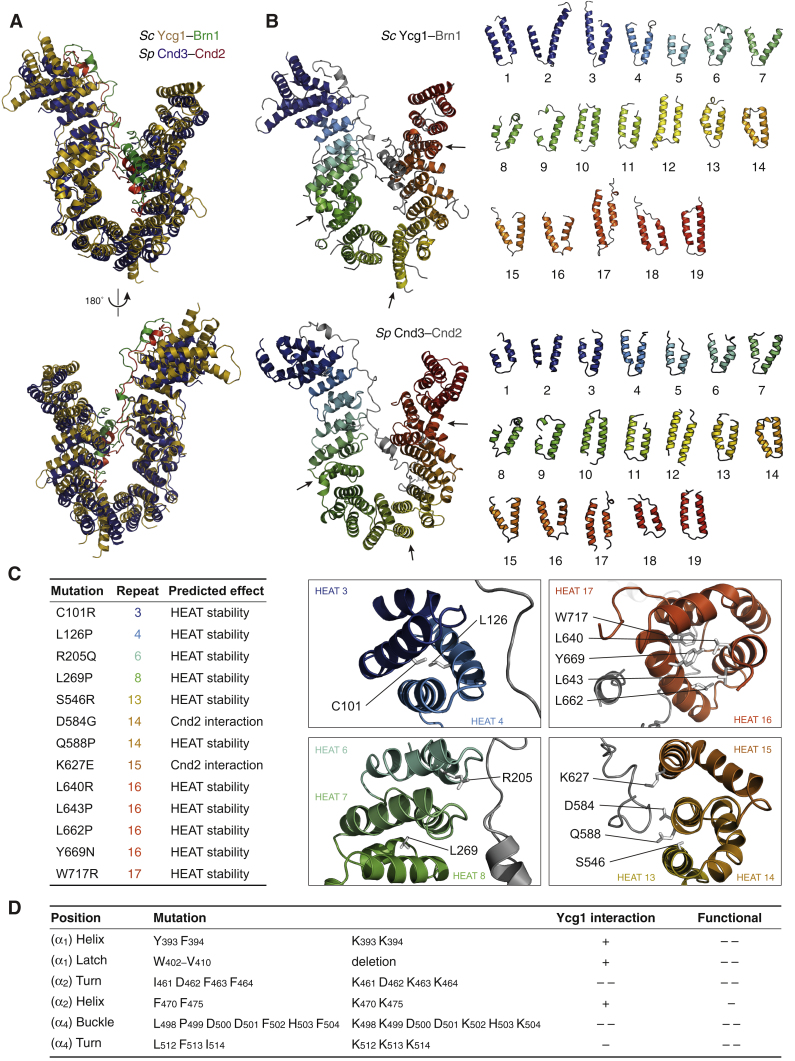
Figure S2.
Comparison of Sp Cnd3–Cnd2 and Sc Ycg1–Brn1 Crystal Structures, Related to Figure 2
(A) Structural alignment of Sp Cnd3–Cnd2 (blue, red) and Sc Ycg1–Brn1 (yellow, green) over all Cα atoms (RMSD 2.53 Å over 715 Cα).
(B) Cartoon representation of the 19 HEAT repeats of Sc Ycg1–Brn1 (top) or of Sp Cnd3–Cnd2 (bottom). Arrows indicate positions of irregularities between canonical HEAT-repeat stretches.
(C) Positions of point mutations in Cnd3 that cause temperature or DNA damage sensitivity in Sp (Petrova et al., 2013, Xu et al., 2015) and their predicted effect based on the Cnd3–Cnd2 structure.
(D) Summary of the effects of previously identified Sc Brn1 mutations on the co-immunoprecipitation of Ycg1 from yeast cell extracts (+ no effect, – reduced Ycg1 co-immunoprecipitation, – – strongly reduced Ycg1 copurification) and on the ability of Brn1 mutant proteins to complement the deletion of the endogenous BRN1 gene (– reduced growth, – – no growth) (Piazza et al., 2014).