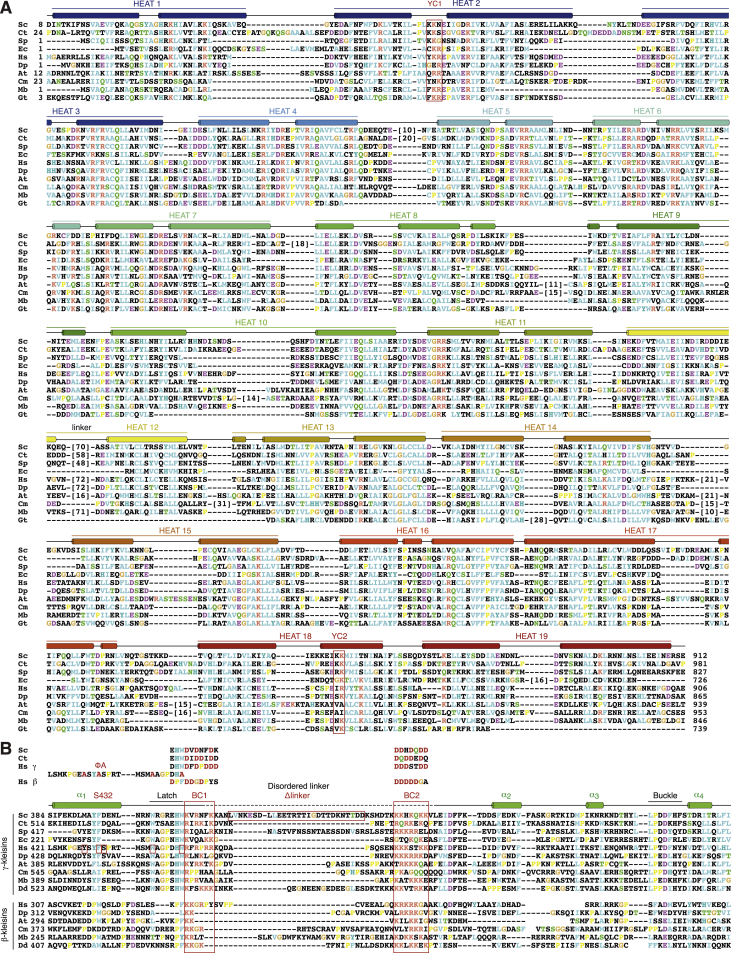
Figure S3.
Multi-sequence Alignment of Selected Condensin HEAT-Repeat and Kleisin Sequences, Related to Figures 2 and 3
(A) Sequence alignment of four yeast (Sc Saccharomyces cerevisiae, Ct Chaetomium thermophilum, Sp Schizosaccharomyces pombe, Ec Encephalitozoon cuniculi), two animal (Hs Homo sapiens, Dp Daphnia pulex), two plant (At Arabidopsis thaliana, Cm Cyanidioschyzon merolae) and two protist (Mb Monosiga brevicollis, Gt Guillardia theta) species selected from an alignment of sequences from 35 divergent species. Secondary structure elements are highlighted based on the Sc Ycg1–Brn1 structure. Sites of mutations in Sc Ycg1 that abolish DNA binding (YC1 and YC2) are highlighted by red boxes.
(B) Sequence alignments of α- (cohesin) and γ- (condensin) kleisins of four yeast and of α-, β- (condensin II), and γ- (condensin I) kleisins of two animal, two plant, and two protist (Dd Dictyostelium discoideum) species selected from an alignment of sequences from 35 divergent species as in (A). Note that yeast genomes encode no β-kleisin subunit. Secondary structure elements are highlighted based on the Sc Ycg1–Brn1 structure. DNA binding site mutations (BC1 and BC2), mutations of hydrophobic latch residues (ΦA), the phosphorylated serine residue in NCAPH (S432), and the region that was deleted in the Sc Brn1Δlinker (short loop) construct are highlighted by red boxes.