Figure S7.
DNA Entrapment by the Brn1 Loop is Required for High-Affinity DNA Interaction, Related to Figure 6
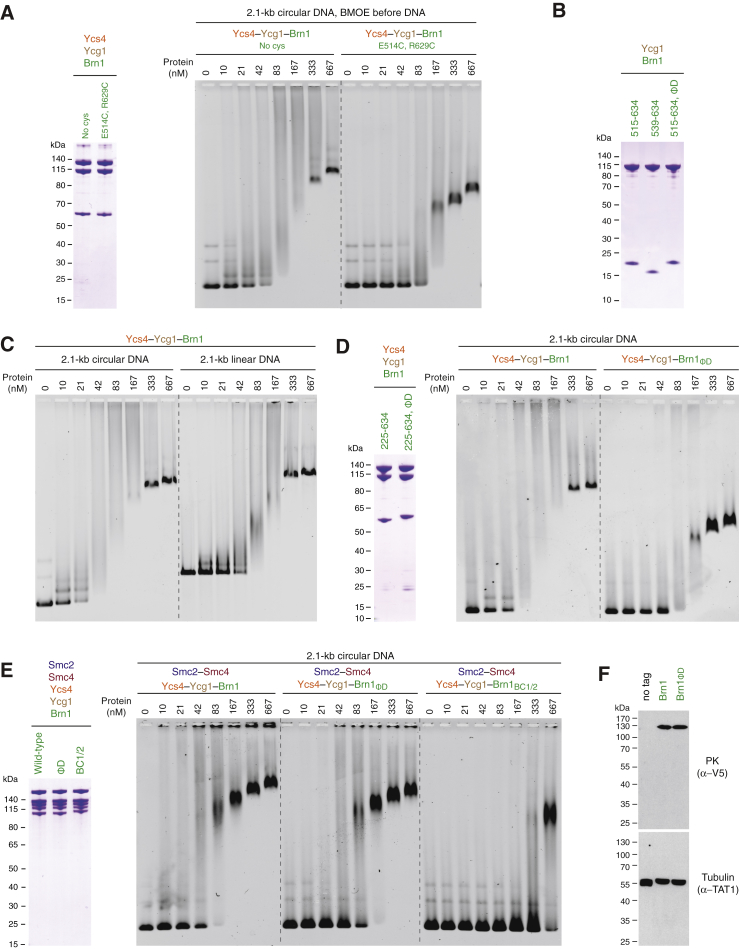
(A) Analysis of copurified Ct Ycs43-1222–Ycg124-1006–His6-TEV-Brn1225-634 complexes without (no cys) or with an additional cysteine pair (Brn1E514C, R629C) engineered into the latch and buckle segments of Brn1 with a 3C protease site following Brn1 residue P549. Complexes were incubated with BMOE crosslinker before analysis by SDS-PAGE and Coomassie staining (left panel) or addition of a 2.1-kb circular DNA (10 nM), followed by EMSA and EtBr staining. Note the differences in the fraction of non-shifted DNA for each protein concentration.
(B) Ct Ycg124-1006– His6-TEV-Brn1 complexes used for EMSA in analyzed by SDS-PAGE and Coomassie staining (see Figures 6B and 6C).
(C) EMSA analysis of the binding of the copurified Ct Ycs43-1222–Ycg124-1006–His6-TEV-Brn1225-634 complex to a 2.1-kb circular DNA (10 nM) or the same DNA linearized by restriction cleavage with XmnI (10 nM).
(D) Analysis of copurified Ct Ycs43-1222–Ycg124-1006–His6-TEV-Brn1225-634 complexes as wild-type or Brn1ΦD latch mutant versions (Brn1L521D, F524D, W532D, W538D) by SDS-PAGE and Coomassie staining (left panel). Protein complexes were used for EMSA with a 2.1-kb circular DNA (10 nM) substrate (right panel).
(E) Analysis of Sc condensin holocomplexes (Smc2–Smc4-StrepII3–Ycs4–Ycg1–Brn1-His12-HA3) as wild-type, Brn1ΦD latch mutant (Brn1L521D, F524D, W532D, W538D) or Brn1BC1/2 charge mutant (Brn1K409D, R411D, K414D, K451D, K452D, K454D, K456D, K457D) versions by SDS-PAGE and Coomassie staining (left panel). Protein complexes were used for EMSA with a 2.1-kb circular DNA (10 nM) substrate (right panel).
(F) Brn1 expression levels of yeast strains C4237, C4239 and C4895 (see Figure 7C) analyzed by western blotting of whole cell lysates against the PK6 tag on Brn1 and α-tubulin as loading control.