Figure 5.
Contacts between Polycomb-Bound Regions Are Disrupted during Neural Differentiation
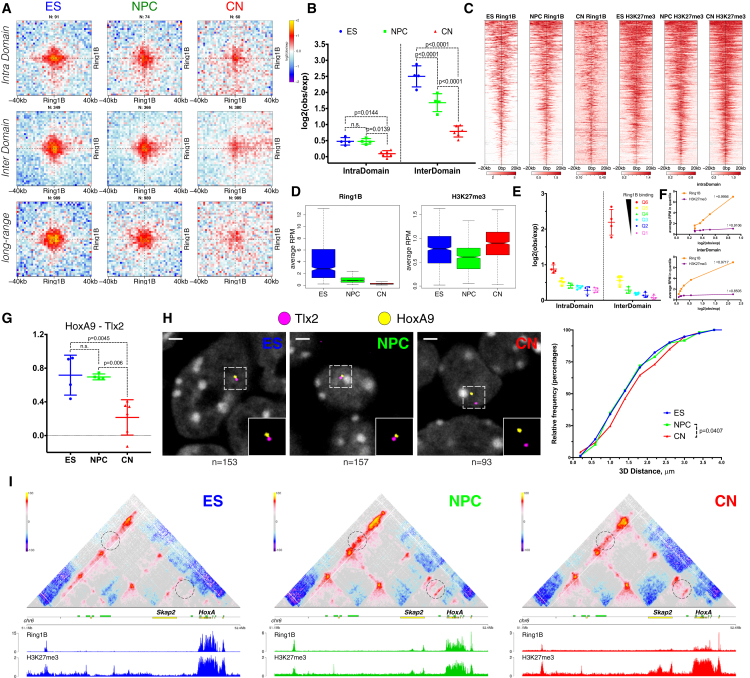
(A and B) Aggregate Hi-C maps and quantification between pairs of regions bound by Ring1B in all three cell types. n refers to the number of pairs examined. Data are represented as a scatter dot plot showing the mean ± SD. Statistical significance is calculated using two-way ANOVA with Tukey’s test.
(C and D) Heat maps and quantification showing the ChIP-seq signal enrichment centered on common Ring1B sites. Note the decrease in Ring1B binding, with no accompanying change in H3K27me3.
(E) Quantification of the contact strength between pairs of the top 3,000 Ring1B binding sites in ESs divided into 6 quantiles based on Ring1B enrichment.
(F) Plots of the average contact enrichment versus the average ChIP enrichment in ESs for either Ring1B or H3K27me3 in each quantile. Shown is also the Pearson correlation coefficient.
(G) Quantification of the contact strength between a long-range Polycomb-associated contact (Tlx2 and HoxA9) based on Hi-C data.
(H) Representative 3D-DNA FISH images (z-slice) and quantification showing the increase in distance between Tlx2 and HoxA9 during differentiation. Statistics are calculated using the Kolmogoroc-Smirnov test.
(I) Hi-C contact maps at the HoxA cluster. NPC-specific contact between Skap2 and the HoxA cluster or an upstream Ring1B binding site are highlighted with a dashed circle.