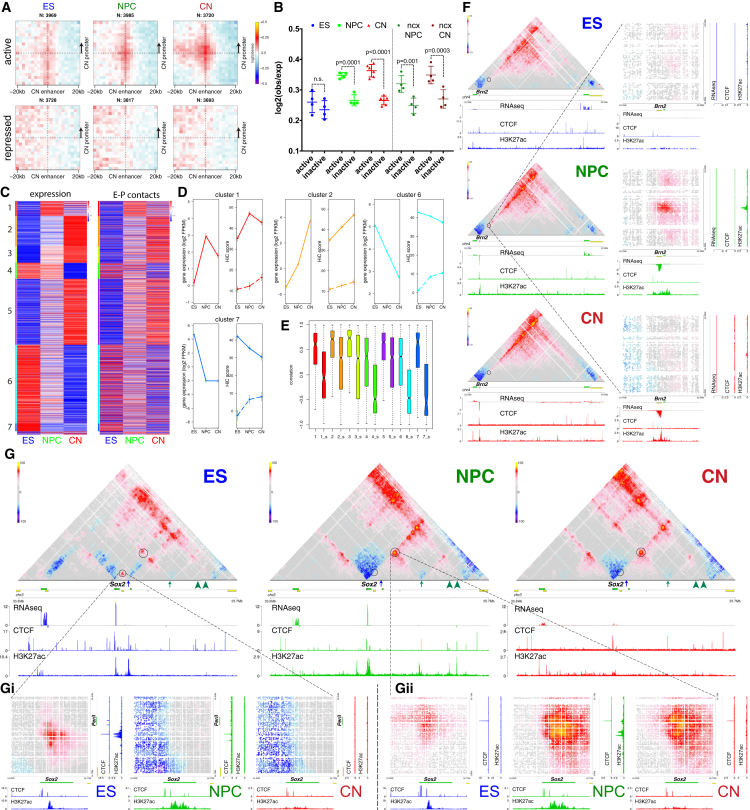
Figure 7.
Enhancer-Promoter Contacts Are Mostly Cell-Type Specific and Are Correlated with Gene Expression
(A and B) Aggregate Hi-C maps and quantification between intra-TAD pairs of enhancers and either active or inactive promoters identified in CNs. Genes were oriented according to the direction of transcription. Data are represented as a scatter dot plot showing the mean ± SD. Statistical significance is calculated using two-way ANOVA with Šídák correction.
(C) Heat map showing Z scores based on either gene expression (fragments per kb of transcript per million mapped reads [FPKM]) or the average E-P interactions per gene. Genes are partitioned using k-means clustering on the RNA expression data across all cell types.
(D) Average expression or average E-P Hi-C score within the specified cluster. Shown are also the average Hi-C scores when the enhancer contact anchor was randomly shuffled within the same TAD (dashed lines). Error bars indicate ± SEM.
(E) Boxplots representing the Pearson correlation coefficient between expression and average E-P interactions per gene, either real or shuffled (shuffled are indicated by the “_s” prefix).
(F) Hi-C contact maps showing ∼2.6-Mb region around Brn2 promoter. Insets show a magnified view of the contact between the Brn2 promoter and an NPC-specific enhancer (dashed circle).
(G) Hi-C contact maps showing ∼2-Mb region at the Sox2 locus. The positions of two putative NPC-specific enhancers are indicated by green arrowheads. Insets showing the interaction between Sox2 promoter and an ES-specific enhancer (blue arrow, Gi); or Sox2 promoter and a known NPC-specific enhancer (green arrow, Gii).