Fig. 2.
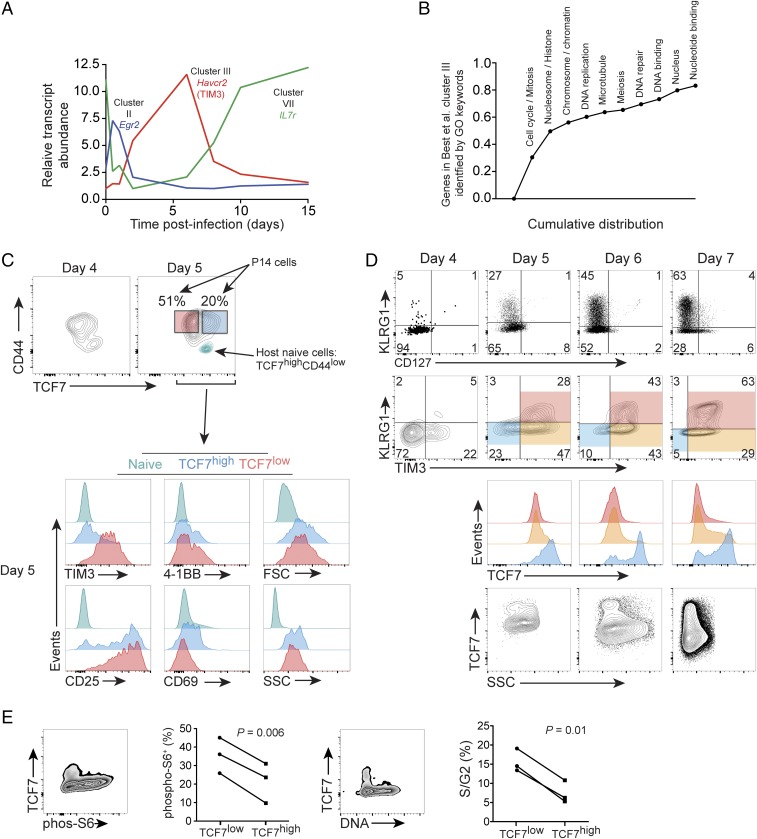
Low TIM3 marks the postinfection TCF7high population. Ten clusters of dynamically regulated genes in a time course of CD8+ T cells responding in vivo to infection were identified by Best et al. (16); primary data are from GSE15907. (A) Kinetics of gene expression of selected transcripts from cluster II, III, and naive/memory-associated cluster VII; cell surface molecule TIM3 (Havcr2) pertains to cluster III. (B) Greater than 80% of genes in cluster III were tagged with one or more of the GO identifiers listed; genes in cluster III were screened for the identifiers in the order listed Left to Right, resulting in a cumulative distribution. (C–E) A total of 1 × 104 P14 cells was adoptively transferred to WT host mice on day −1 and was infected with LCMV-ARM on day 0. (C, Above) Day 4 and day 5 postinfection P14 expression of TCF7 and CD44. For day 5, the naive host CD8+ population (CD44low) from the same sample is plotted in teal for intrasample reference of high TCF7 and low CD44. (Below) The expression of indicated proteins, FSC-A and SSC-A, on day 5 using the color-coded gates established in the day 5 contour plot above. (D) CD127 and TIM3 vs. KLRG1 on total P14 cells from day 4 to 7 postinfection. TCF7 expression within the indicated shaded gates is then plotted below. Total P14 SSC-A vs. TCF7 for indicated days is shown below. Where present, Insets indicate the percentage of the gated population. Note absolute values of immunofluorescence may vary among days, as immunostaining was performed independently each day. Experiments in C and D performed twice with similar results. (E) Gates were established on the P14+KLRG1− adoptively transferred cells, and immunostaining for phospho-S6 or determination of DNA content with DRAQ5 was performed at day 6–7 postinfection. The percent positive for phospho-S6 or in S/G2 of the cell cycle was determined; n = 3, three independent experiments, Student’s paired t test.