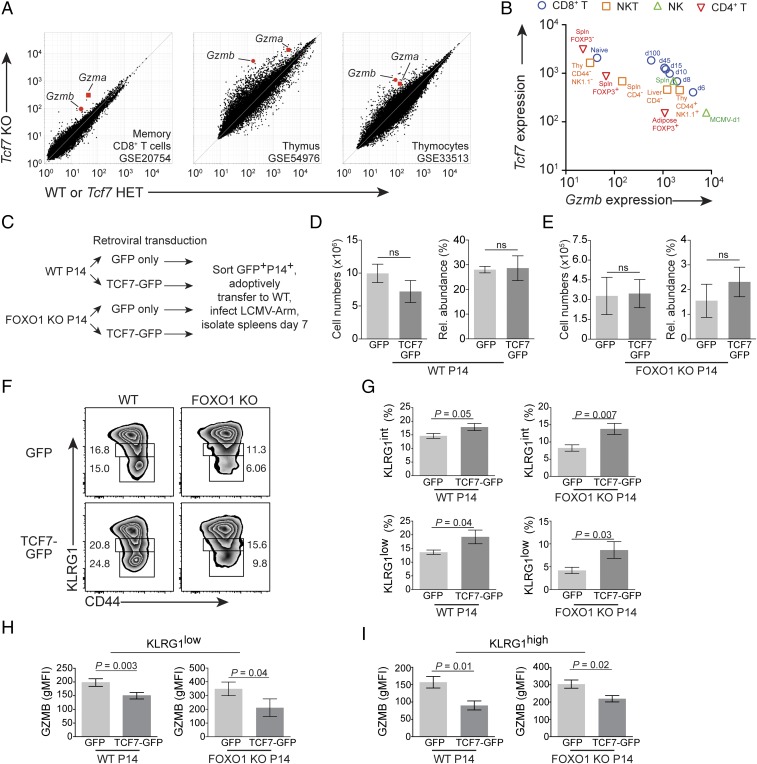
Fig. 4.
Tcf7 forestalls phenotypic terminal differentiation and opposes immune cell Gzmb. (A) Gzma and Gzmb expression in three published microarray studies containing Tcf7 mutant CD8+ T cells. (Left) Peripheral, postinfection memory CD8+ T cells; (Center) thymus; and (Right) mouse DN1 stage thymocytes (Lin− CD44+CD25−KIT+). Inset summarizes cell type and NCBI GEO accession number. (B) Plot of Gzmb vs. Tcf7 gene expression; color/shape indicate cell type. Note further annotation in plot (i.e., CD44, NK1.1), tissue/organ (i.e., adipose, spleen), or infection (i.e., MCMV, day 1 postinfection shown for NK cells responding to MCMV). For CD8+ T cells, d6, d8, d10, d15, d45, and d100 indicate days postinfection with Listeria monocytogenes-OVA. Primary data in B are from GSE15907. (C) Experimental diagram for retroviral transduction. (D–I) WT P14 GFP+-gated and FOXO1 KO P14+GFP+-gated cell numbers and phenotypes following protocol in C. Cell number and relative abundance of (D) WT and (E) FOXO1 KO. Representative KLRG1 immunophenotype with (F) KLRG1low and KLRG1int gates shown, and (G) pooled KLRG1 phenotype data. GZMB expression in (H) KLRG1low and (I) KLRG1high P14 cells. Error bars are SEM; P values are from Student’s unpaired t test. GFP, n = 9; WT GFP-TCF7, n = 7; FOXO1 KO GFP, n = 7; FOXO1 KO GFP-TCF7, n = 6. Retroviral transduction and TCF7-dependent decrease in GZMB were observed three times; pooled data are from three independent experiments plotted in G–I.