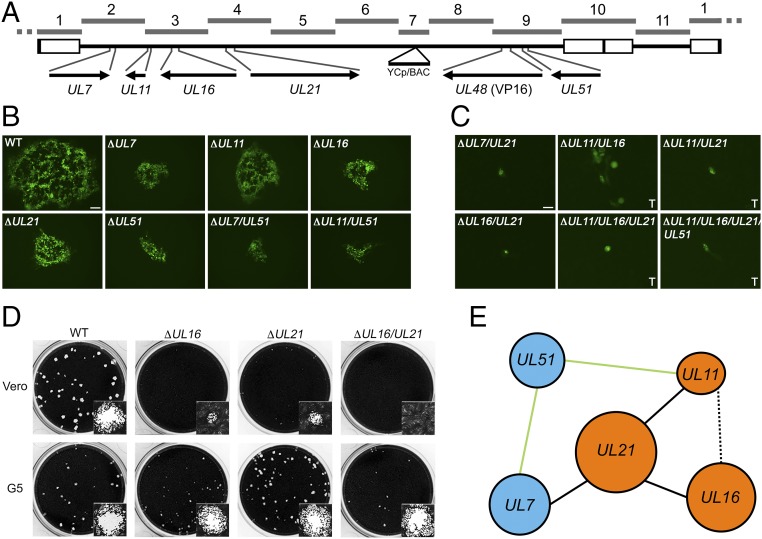
Fig. 5.
Replication of viruses containing combinatorial deletions of five tegument genes. (A) A linear map of the HSV-1 genome shows the locus of each gene targeted for modification and its location in the respective TAR fragments. The UL48 gene encodes the VP16 protein. (B) Recombinant viruses, each containing a different tegument gene deletion or combination of deletions and the VP16Venus fusion, which replicate in Vero cells are shown. Plaques were visualized using wide-field fluorescence light microscopy 3 d postinfection. (Scale bar, 100 μm.) Wild-type is KYAVP16Venus and contains no other gene modifications. (C) KOSYA mutant viruses that did not exhibit productive infection in Vero cells were infected and imaged as indicated in B or recombinant genomes were transfected (marked “T”) into Vero cells and imaged 6 d posttransfection. (Scale bar, 25 μm.) (D) KOSYA viruses containing a deletion of UL16, UL21, or combination thereof were infected at low MOI on Vero and G5 cell monolayers in 12-well trays. Cells were stained with Crystal violet 3 d postinfection to visualize plaque formation. Insets show a single plaque. Stained plaques were imaged using the CCD (charge-coupled device) camera of a Gel Doc XR+ (Bio Rad) and insets show a representative single plaque imaged with the brightfield settings of a Zoe Fluorescent Cell Imager (Bio Rad) with a 20× objective lens. (E) Each of the genes analyzed in this study are shown and blue or orange is used to highlight previously discovered physically interacting complexes (55). Solid black lines indicate essential genetic pairs (i.e., UL21 and UL7), whereas solid green lines indicate nonessential pairs (i.e., UL51 and UL11). A dashed black line represents a pair, UL11 and UL16, resulting in severely diminished, but partial, biological activity.