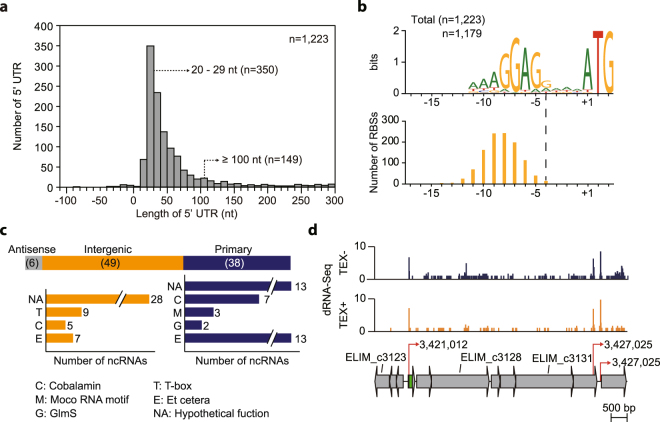
Figure 5.
Genome-scale analysis of 5′UTR and functional prediction of regulatory ncRNAs. (a) Distribution of 1,223 5′UTRs of E. limosum, with a major peak at length 20–29 nt. (b) The conserved ribosome binding site (RBS) motif (GGAGR) for protein coding genes in the 5′UTRs is longer than 8 nt. The bottom panel shows conservation of each motif of RBS. (c) Classification of ncRNAs using antisense, intergenic, and primary TSSs, according to their locations. The functions of the classified RNAs were predicted using the Rfam database. (d) Moco-RNA motif containing ncRNA, which was identified by dRNA-Seq, was located upstream of the hydrogenase complex at genome position 3,421,126, which was confirmed via the Rfam database.