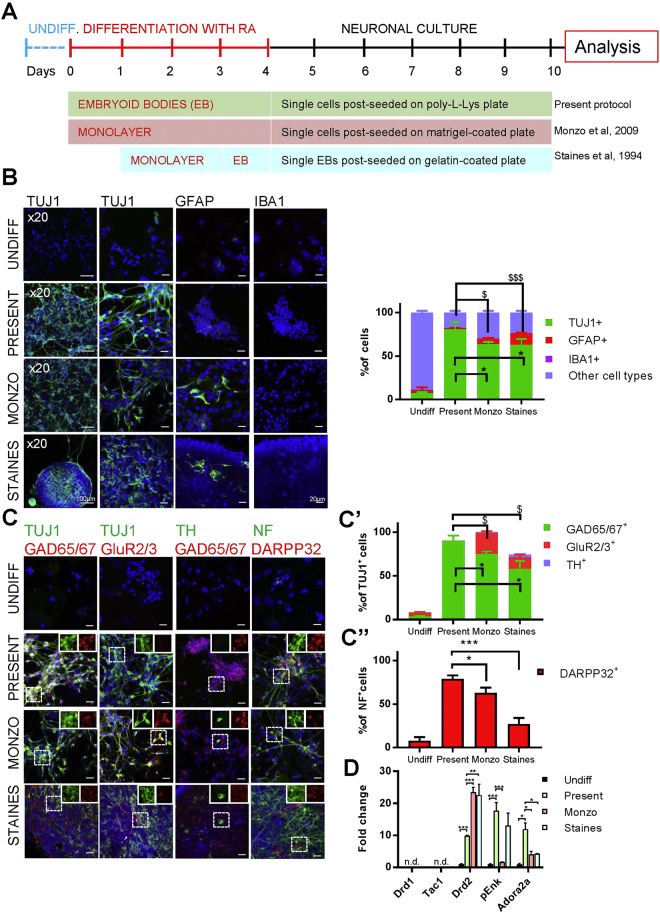
Figure 5.
Differentiation of EC cells using distinct protocols. (A) Experimental design for neuronal differentiation of EC cells using embryoid bodies (EB), monolayers, and a mixture of EB and monolayers, according to published protocols (Staines et al., Monzo et al., see Main text for refs). (B) Generation of neurons, astrocytes and microglial cells was assessed using immunocytochemistry for three distinct protocols (Staines, Monzo, present protocol; see examples on left panels and quantifications on the right panel). *p < 0.05 as compared to number of TUJ1+ cells generated by present protocol. $p < 0.05, $$$p < 0.001 as compared to number of GFAP+ cells obtained in present protocol. (C) Immunofluorescence analysis of neuronal populations: inhibitory (GAD65/67+), excitatory (GluR2/3+), dopaminergic (TH+) and MSN (DARPP32+) neurons. (C’) Quantification of different types of TUJ1+ neurons and (C”) NF+ MSNs obtained with distinct protocols. *p < 0.05 as compared to number of GAD65/67+ neurons obtained by present protocol. $p < 0.05 as compared to number of GluR2/3+ neurons generated by present protocol. (D) mRNA expression level of striatonigral and striatopallidal markers quantified by RT-qPCR for each protocol. Three independent samples were used for each experimental condition. The expression of each marker is plotted as fold change with respect to undifferentiated cells. ***p < 0.001, *p < 0.05. n.d., not detected. Throughout the figure, graph bars represent means ± s.e.m. Scale bars: 100 µm (x20 panels), 20 µm (others).