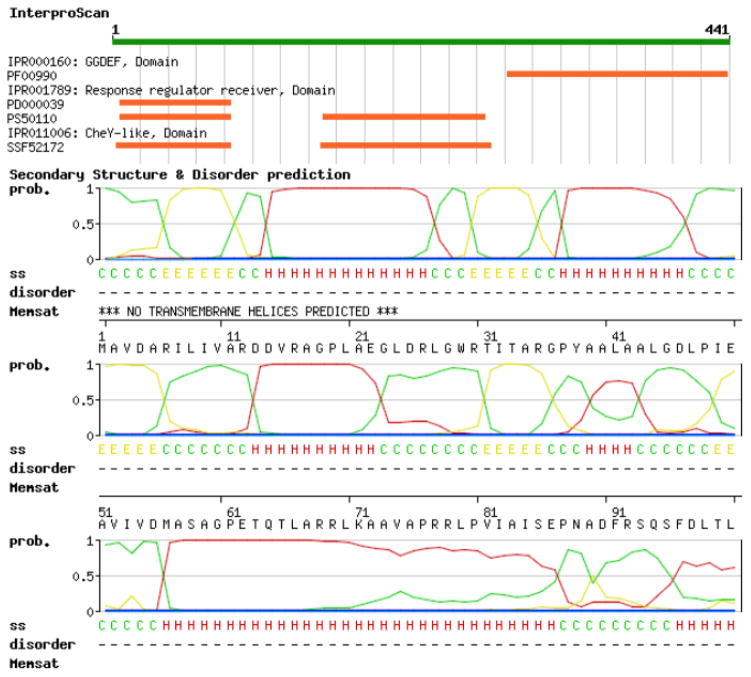
Fig. 2.
SWISS-MODEL Workspace target sequence feature annotation. To predict functional and structural features of the target proteins, several annotation tools are available on the SWISS-MODEL Workspace. In this example, the C. crescentus PopA protein (represented as a green bar on the top) is predicted to contain a C-terminal GGDEF domain and two N-terminal receiver domains. The likelihood (between 0 and 1, where 1 means highest probability) of the occurrence of secondary structure elements are depicted as curves (red for alpha-helices, yellow for β-strands and green for coiled regions). Prediction of disordered regions and transmembrane domains is also available. In particular, for PopA neither intrinsically unstructured regions nor portions of the protein spanning the membrane are detected.