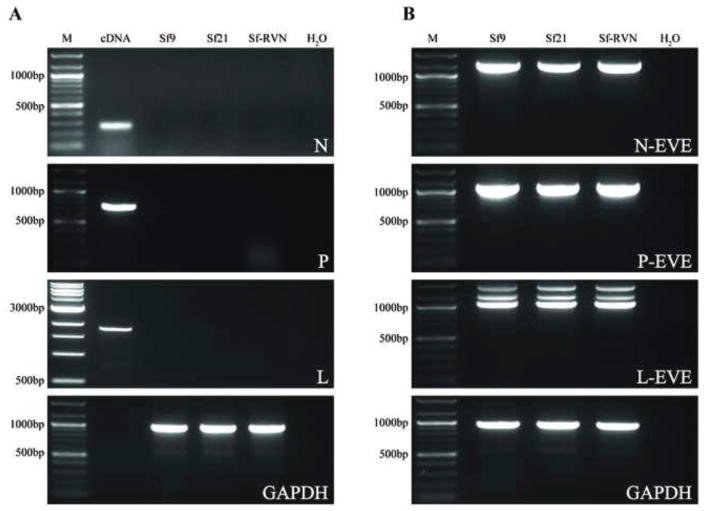
Fig. 3.
Genomic integration of Sf-rhabdovirus and Sf-rhabdovirus-related EVE sequences. Genomic DNAs were isolated from Sf9, Sf21, and Sf-RVN cells, as described previously (Geisler and Jarvis, 2016) and used for PCRs with (A) Sf-rhabdovirus N-, P-, or L-specific or (B) Sf-rhabdovirus N-, P-, or L-related EVE-specific primers, as described in Materials and methods. PCRs with no template (H2O) were used as negative controls and PCRs with the indicated templates and Sf glyceraldehyde phosphate dehydrogenase (GAPDH)-specific primers were used as positive controls. PCRs with total Sf cellular cDNA as the template and the Sf-rhabdovirus N-, P-, and L-specific primers were used as an additional positive control in panel A. Note we did not include a cDNA template control in the GAPDH primer set, so no sample was loaded into the lane marked cDNA in the lowermost panel of A. The lane marked M shows the 100-bp or 1000-bp markers, with selected sizes indicated on the left.