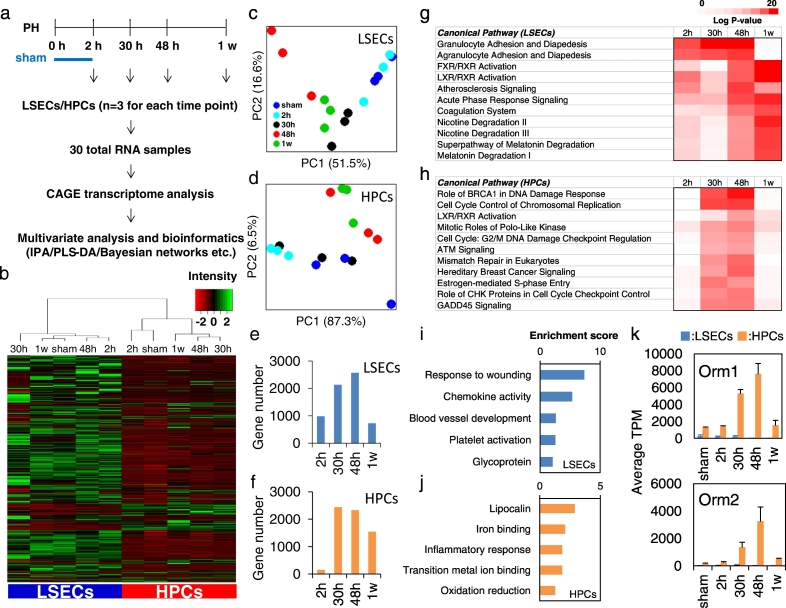
Fig. 2.
Transcriptional profiling of LSECs and HPCs during liver regeneration. (a) Schematic overview of transcriptome experimental procedures (n = 3). (b) Hierarchical clustering with Ward's method of 16,499 genes measured by CAGE technology in LSECs and HPCs. The mean average from three biological replicates at each time point was shown. Low expression tags were deleted and the tags that were expressed at least at one time point in either LSECs or HPCs were kept with the expression cutoff at 1 tags per million (TPM). PCA score plots of differentially expressed genes in the process of liver regeneration for (c) LSECs and (d) HPCs. Numbers of significantly differentially expressed genes compared with the sham control at each time point in (e) LSECs and (f) HPCs. Top canonical pathways associated with significantly differentially expressed genes during liver regeneration in (g) LSECs and (h) HPCs in IPA program. Functional annotations identified using DAVID software with top (i) LSEC and (j) HPC genes selected by evaluating the relative contribution of each gene to differential gene expression during the process of liver regeneration in PLS-DA modeling. (k) Expression of Orm1 and Orm2 in LSECs and HPCs during liver regeneration measured by CAGE analysis.