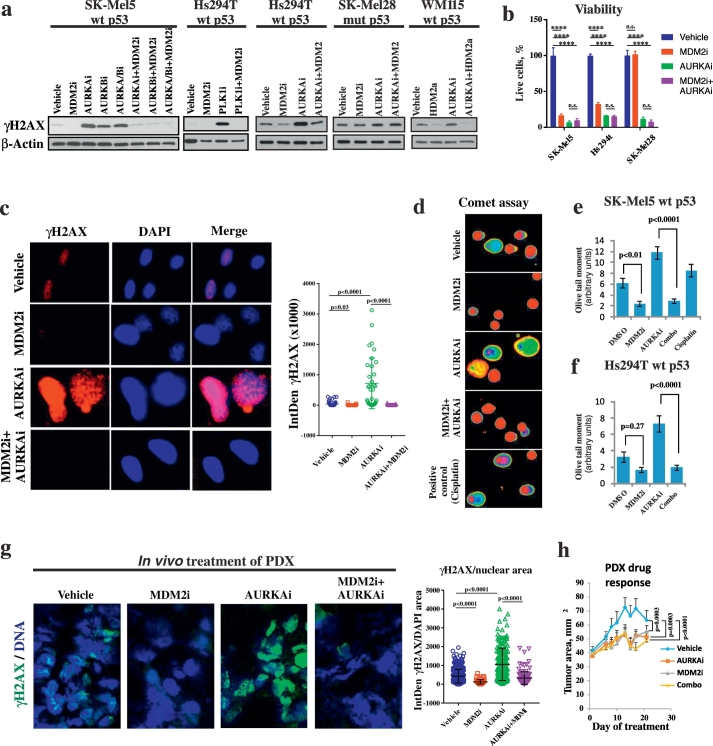
Fig. 1.
MDM2i counteracts DNA damage induced by mitotic inhibitors in p53WT cells. (a) Western Blot analysis of DNA damage marker γH2AX in the indicated melanoma cells after 3 days of treatment with 10 μM nutlin-3a (MDM2i) or 1 μM idasanutlin (HDM2i) in combination with 1 μM alisertib (AURKAi), 1 μM borasertib (AURKBi), 5 μM danusertib (AURKA/Bi) or 5 nM volasertib (PLK1i). (b) Count of viable cells from indicated cell lines after 5 days treatment with vehicle, 1 μM AURKAi, 10 μM MDM2i, or the combination of both drugs. Mean ± SD from 2 biological replicates and results of ANOVA with Tukey's comparison are shown. (c) Immunofluorescent staining with γH2AX-specific antibody in SK-Mel5 cells treated as in (a). Images were quantified using ImageJ and DNA damage was measured as IntDen of γH2AX within the nucleus area. Five fields were quantified for each condition for the total of 56 cells in vehicle, 33 in MDM2i, 34 cells in AURKAi, and 23 in AURKAi + MDM2i-treated group. Raw values from individual cells, mean, SEM and Kruskal Wallis test results are shown. (d–f) Single cell electrophoresis (comet assay) in SK-Mel5 cells. Representative pseudo-colored images of fluorescent-labeled DNA after analysis using Comet Score software (d) and quantitative results (mean olive tail moment in individual cells ± SEM, e-f) are shown. About 200 cells were counted in each treatment group The Mann-Whitney test was used for statistical evaluation. Treatment with cisplatin (10 μM, 48 h) was used as a positive control. (g) Patient-derived human melanoma tissue was implanted SC into nude mice. When tumors reached the size of ~ 100 mm3 mice received daily treatments with nutlin-3a (200 mg/kg), MLN8237 (30 mg/kg), a combination of both, or vehicle control for 3 weeks. Representative images after IF staining for γH2AX. The amount of γH2AX per nuclear area was quantified using ImageJ across several fields. In total, 368 cells in vehicle, 59 in MDM2i, 176 cells in AURKAi, and 131 in AURKAi + MDM2i-treated group were quantified. Kruskal Wallis statistics are shown. (h) Tumor progression over time for tumors analyzed in (g). Tumor area (length x width) was measured every 3–4 days. Average tumor area ± SD is shown. N = 8 in combination groups and n = 10 in other treatment groups. Mixed-effects statistical model was used to assess group differences in tumor area over days. All in vitro experiments were repeated at least 3 times with consistent results and results from a representative experiment are shown. ****p < 0.0001, n.s. – not significant (p > 0.05).