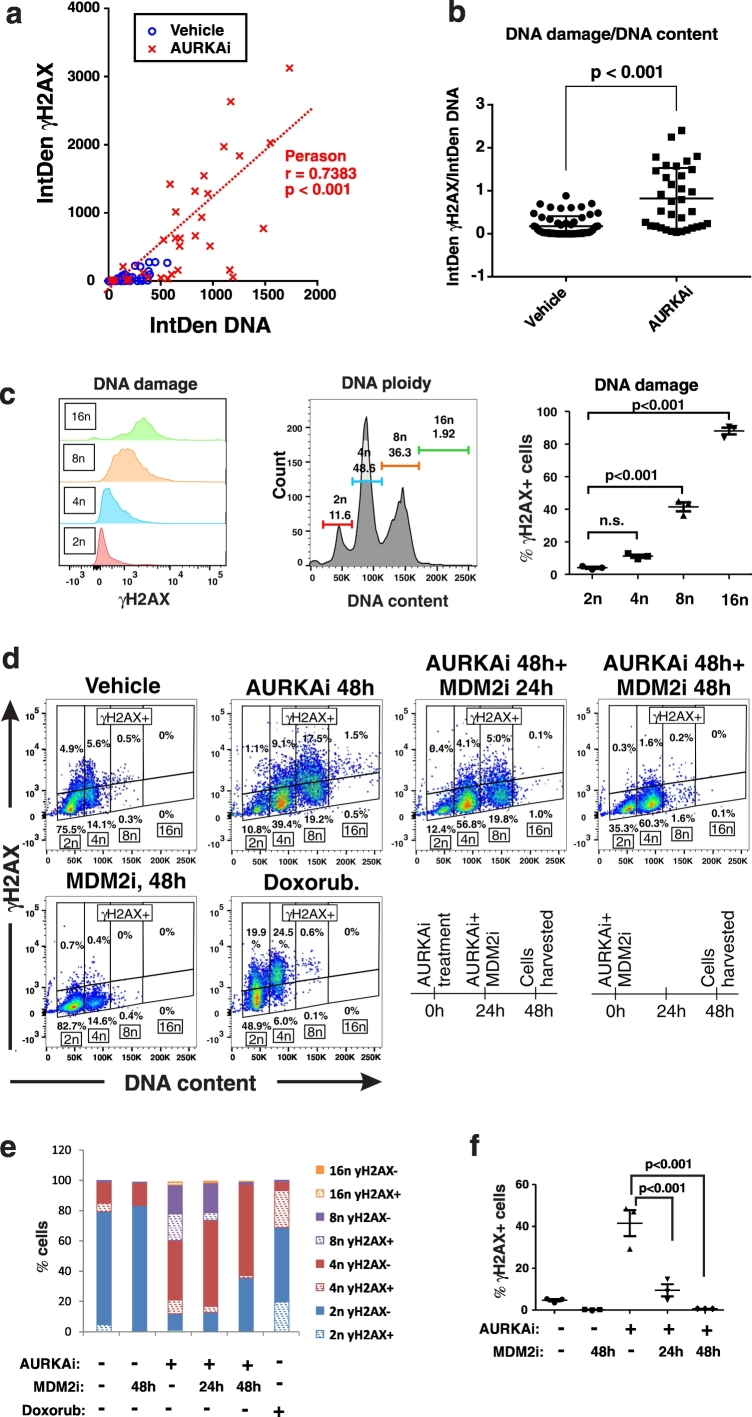
Fig. 3.
AURKAi-induced DNA damage is associated with polyploidy. (a) The scatter plot shows distribution of individual vehicle or AURKAi (1 μM alisertib, 3 days) treated SK-Mel5 cells based on their DNA content (x-axis) and DNA damage (y-axis). These parameters were derived from the ImageJ analysis of IF staining shown on Fig. 1b. DNA content is measured as integrated density of DAPI staining (IntDen DNA), and DNA damage is measured by IntDen of γH2AX staining within the same nucleus. Five fields were quantified for each condition for the total of 56 cells in vehicle and 34 cells in the AURKAi-treated group. The results of Pearson correlation test are shown. (b) DNA damage adjusted for DNA content in vehicle and AURKAi-treated SK-Mel5 cells shown in (a). Individual values, means, SEM, and Mann-Whitney test results are shown. (c) Expression of DNA damage marker γH2AX in SK-Mel5 cells with 2n, 4n, 8n, and > 8n ploidy after treatment with 1 μM alisertib. Over 10,000 cells were analyzed by flow cytometry. Values from 3 independent experiments are shown as a dot plot on the right. Statistical comparison was performed using ANOVA with Dunnett's post-test (d) Flow cytometry analysis of DNA damage and DNA ploidy in SK-Mel5 cells treated with vehicle or 1 μM alisertib (AURKAi) for 2 days. Groups of AURKAi-treated were also co-treated with 10 μM of nutlin-3a (MDM2i) for the full duration of 2 day treatment (AURKAi 48 h + MDM2 48 h) or only for the last 24 h of treatment (AURKAi 48 h + MDM2 24 h). Doxorubicin treatment (20 nM, 2 days) was used as positive control for DNA damage induction. A schematic depicting treatment schedule is shown at the bottom. (e) Distribution of SK-Mel-5 cells in the gates set based on cell ploidy (2n, 4n, 8n, 16n) and γH2AX expression (γH2AX- or γH2AX +) as shown in (d). (f)SK-Mel5 cells were treated as described in (d) and analyzed for γH2AX levels by flow cytometry. Dot plot shows data from 3 biological replicates, as well as mean ± SD and ANOVA with Tukey's post-test results.