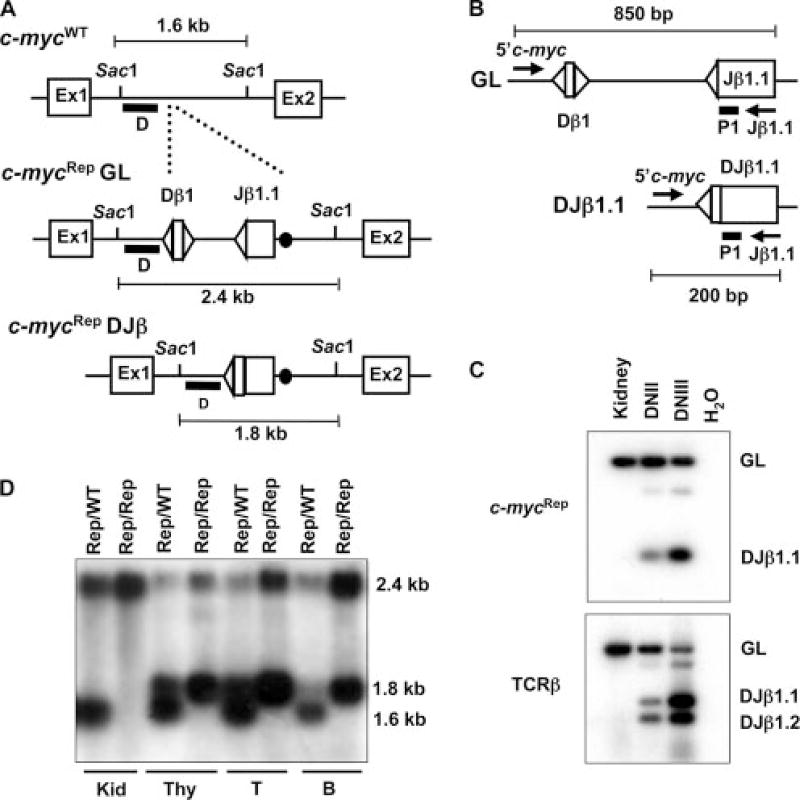
FIGURE 1.
Generation and analysis of c-mycRep mice. A, Schematic diagrams of the WT c-myc locus (WT), the c-myc locus with the inserted germline Dβ1 and Jβ1.1 segments (c-mycRep). The RSs are depicted as triangles and the loxP site as a black circle. Open boxes depict the relative locations of the three c-myc exons. The relative location of the SacI sites and the sizes of the SacI-digested genomic fragments for WT, c-mycRep germline (GL), and c-mycRep DJβ alleles are indicated. ■, The location of the 5′Vβ14, DJβ1, and 3′Vβ14 probes. B, Schematic diagrams of the GL and Dβ to Jβ rearranged (DJβ) c-mycRep alleles. The relative location of the 5′c-myc and Jβ1.1 primers are indicated with arrows. ■, The location of the P1 probe. The sizes of the PCR products for c-mycRep GL and c-mycRep DJβ alleles are indicated. C, PCR analysis of c-mycRep Dβ to Jβ rearrangements, endogenous Dβ to Jβ rearrangements, and endogenous Vβ14 to DJβ rearrangements using the 5′c-myc/Jβ1.1. 5′Dβ1/P2, and Vβ14/P2 primer sets on genomic DNA isolated from Vβ14Rep/ω kidneys and sort-purified DNII or DNIII thymocytes. Products corresponding GL and rearranged (DJβ1.1) c-mycRep alleles, as well as endogenous TCRβ rearrangements, are indicated. D, Southern blot analysis of c-mycRep Dβ to Jβ rearrangements using probe D on SacI digested genomic DNA isolated from c-mycRep/WT or c-mycRep/Rep kidney, thymus, T cells, or B cells. Restriction fragments representing GL and rearranged (DJβ1.1) c-mycRep alleles, as well as endogenous c-myc alleles, as indicated.