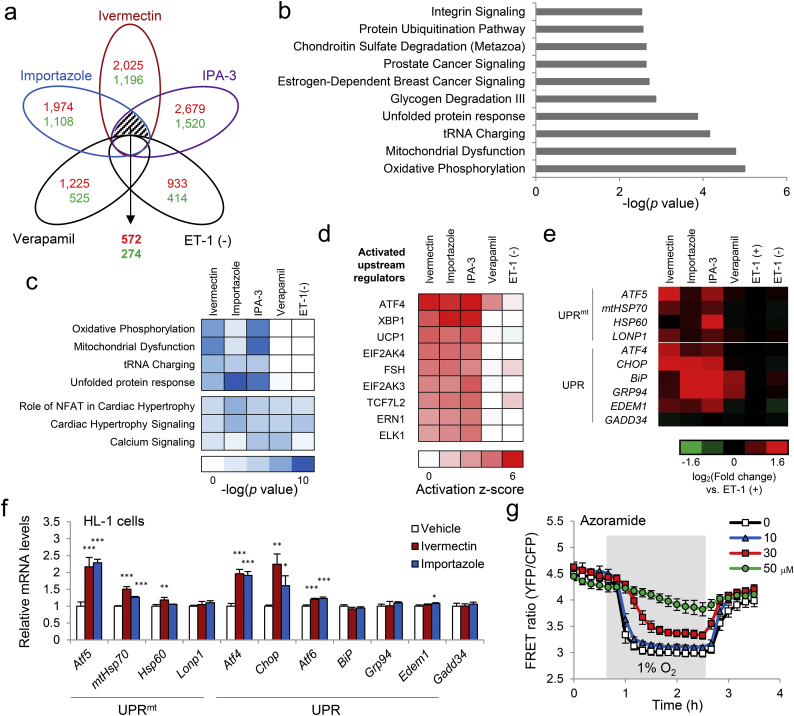
Fig. 7.
Induction of mitochondrial unfolded protein response (UPR) following mitochondrial ATP modulator treatment in human iPSC-derived cardiomyocytes. (a) Venn diagram of differentially expressed genes identified from AmpliSeq data in iPSC-derived cardiomyocytes untreated [ET-1 (−)], or treated with 3 μM ivermectin, 10 μM importazole, 10 μM IPA-3, or 1 μM verapamil in the presence of 10 nM ET-1 for 18 h (n = 4 biologically independent samples for each condition, fold change ≥ 1.5, p < 0.05 vs. ET-1 treatment). Red, the number of upregulated genes; green, the number of downregulated genes for each treatment. (b) Pathways enriched in commonly overlapped genes only among ivermectin, importazole, and IPA-3 were identified by Ingenuity Pathway Analysis (IPA). (c) The gene expression signature was subjected to the pathway association analysis (IPA pathways) and hierarchical cluster analysis. The heatmap shows representative IPA pathways enriched in two clusters (ivermectin, importazole, and IPA-3 groups, and all five groups). (d) Upstream analysis was performed using IPA and was subjected to hierarchical cluster analysis. The heatmap shows the top nine upstream regulators clustered into ivermectin, importazole, and IPA-3 groups. (e) Heatmap of AmpliSeq data of genes involved in mitochondrial UPR (UPRmt) and UPR in iPSC cardiomyocytes treated with the indicated compounds as in A. Red, upregulated genes; green, downregulated genes; fold change (log2) ≥ 1.6. (f) Relative mRNA levels of genes involved in UPRmt and UPR in HL-1 cardiomyocytes treated with the indicated compounds. Data are presented as means ± SD (n = 3 biologically independent samples). ⁎p < 0.05, ⁎⁎p < 0.01, ⁎⁎⁎p < 0.001 vs. vehicle by one-way ANOVA with Dunnett's multiple-comparison test. (g) FRET ratio of mito-ATeam stable HL-1 cardiomyocytes treated with azoramide. Data are presented as means ± SD (n = 3 biologically independent samples).