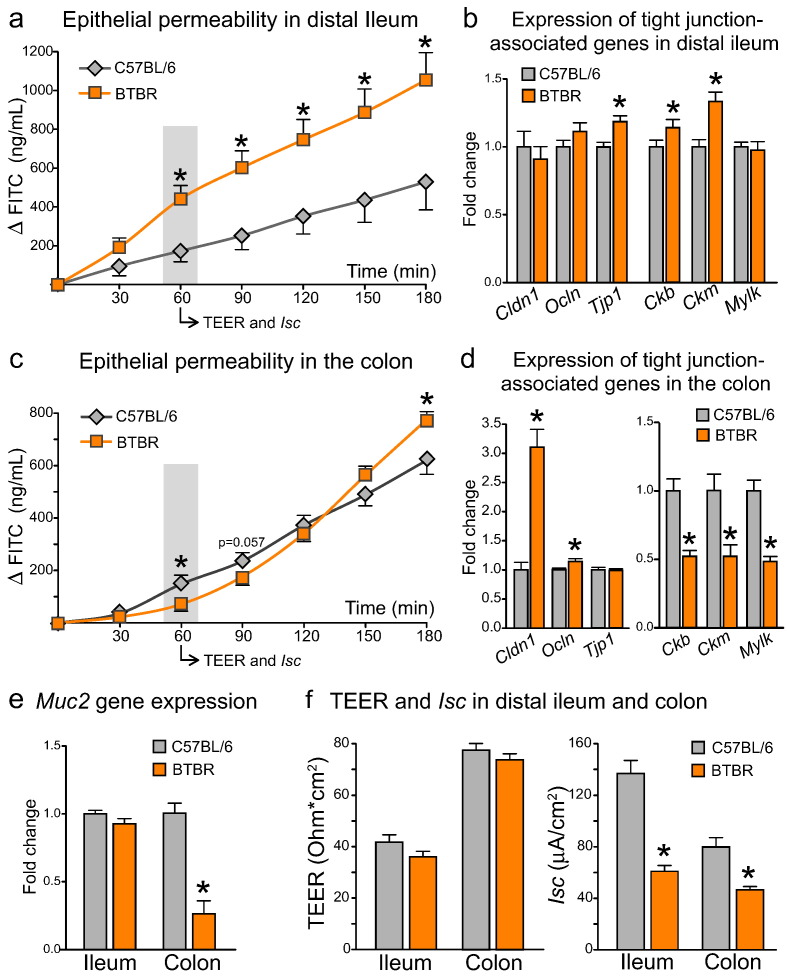
Fig. 4.
Differential changes to epithelial permeability and tight junction-associated gene expression in small and large intestine of BTBR mice (a) In distal ileum, BTBR mice showed a marked increase in FITC transepithelial permeability, with no substantial changes to either tight junction-associated genes (b) or mucin2 gene expression (e). (c) In the colon of BTBR mice, FITC permeability was increased in the end, but decreased in the beginning of the incubation period. The biphasic response of FITC flux was coupled with an up-regulation of tight junction protein genes, but a down-regulation of creatine kinase and mucin2 genes (d–e). (f) TEER was not changed, while Isc was substantially decreased in the BTBR intestine. *p < 0.05 for BTBR vs. C57BL/6 group. Statistical analysis: data are presented as mean + SEM, n = 10 in either group. FITC permeability in distal ileum: F(6;84) = 63.339, p < 0.0005 for the effect of Time, F(1;14) = 7.090, p = 0.019 for the effect of Genotype, F(6;84) = 7.177, p = 0.015 for the Time × Genotype interaction. FITC permeability in the colon: F(6;102) = 383.858, p < 0.0005 for the effect of Time, F(1;17) = 0.061, p = 0.808 for the effect of Genotype, F(6;102) = 9.198, p = 0.002 for the Time × Genotype interaction. Datasets were analyzed with mixed-design ANOVA with Genotype as an independent factor and Time as a repeated measure. Mean values in each time point were further compared between groups with unpaired t-test. t(14) = 1.582, p = 0.136 for TEER; t(14) = 6.862, p < 0.0005 for Isc; t(18) = 0.624, p = 0.541 for Cldn1, t(18) = − 1.429, p = 0.170 for Ocln, t(18) = − 2.768, p = 0.013 for Tjp1, t(15) = − 3.292, p = 0.005 for Ckb, t(16) = − 3.731, p = 0.002 for Ckm, t(15) = 0.217, p = 0.831 for Mylk, t(17) = 1.278, p = 0.218 for Muc2 gene expression in distal ileum, unpaired t-test. t(18) = 1.101, p = 0.285 for TEER; t(18) = 4.407, p = 0.001 for Isc; t(18) = − 6.319, p < 0.0005 for Cldn1, t(18) = − 2.273, p = 0.035 for Ocln, t(18) = 0.104, p = 0.918 for Tjp1, t(17) = 4.791, p < 0.0005 for Ckb, t(17) = 3.255, p = 0.005 for Ckm, t(17) = 5.751, p < 0.0005 for Mylk, t(17) = 6.261, p < 0.0005 for Muc2 gene expression in the colon, unpaired t-test.