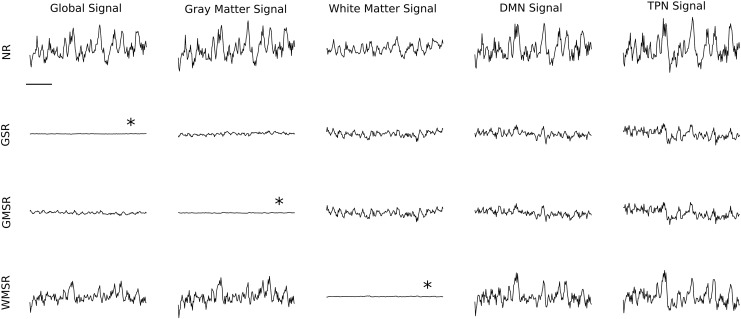
FIG. 1.
Effects of tissue-specific regression on BOLD signal amplitude. Data from a single subject (New Haven dataset). Averaged BOLD signal time courses in the (left to right) global signal (whole brain), gray matter signal, white matter signal, DMN signal, and TPN signal. The BOLD signal time courses shown are NR data before regression (first row), after GSR (second row), after GMSR (third row), and after WMSR (fourth row). The horizontal bar denotes 60 sec. All signals are scaled to represent fractional change (see the Results section), that is, vertical scales are arbitrary, but identical. Asterisks denote that these signals are expected to be flat because these were the regional signals that were regressed. BOLD, blood oxygenation level-dependent; DMN, default mode network; GMSR, gray matter signal regression; GSR, global signal regression; NR, nonregressed; TPN, task-positive network; WMSR, white matter signal regression.