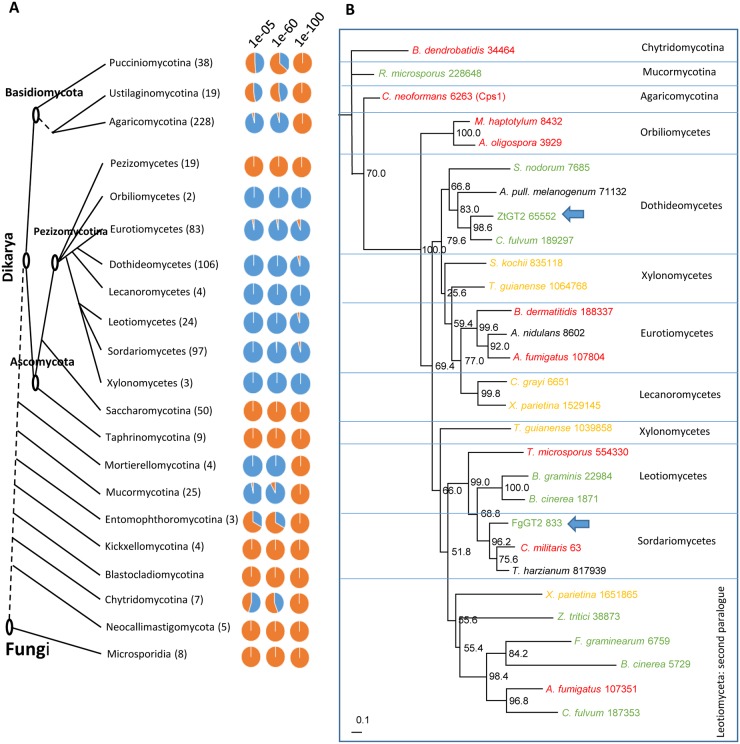
Fig 3. ZtGT2 orthologues show a discontinuous distribution throughout the fungal tree of life but are present in saprophytes, mutualists and pathogens.
(A) Diagrammatic representation of the fungal tree of life adapted from the JGI MycoCosm genome portal (http://genome.jgi.doe.gov/programs/fungi/index.jsf). Taxonomic tree distances are only representative and do not accurately indicate evolutionary time. Each fungal taxonomic class or sub phylum is shown with brackets highlighting the number of species genomes present in each group analysed. Pie charts then display the number of species genomes which have a at least one similar protein to ZtGT2 at each of the indicated Blastp expect thresholds. Blue shading indicates the number of species with at least one similar proteins. Orange shading indicates the number of species with no similar proteins. (B) Maximum Likelihood phylogenetic tree displaying representative sequence orthologues to ZtGT2 from across the fungal tree of life. Node labels indicate percentage bootstrap support (500 replicates). Where similar genes were detected orthology is inferred by correspondence to the fungal species tree. Species names in green font indicate known plant pathogens, those in red font indicate known animal pathogens and those in yellow indicate those with mutualistic lifestyles. A true saprophytic lifestyle is included in the form of Aspergillus nidulans and other species in black font have currently ambiguous lifestyles. Numbers following species names represent the protein model Id retrievable from the JGI Mycocosm species genome portal.