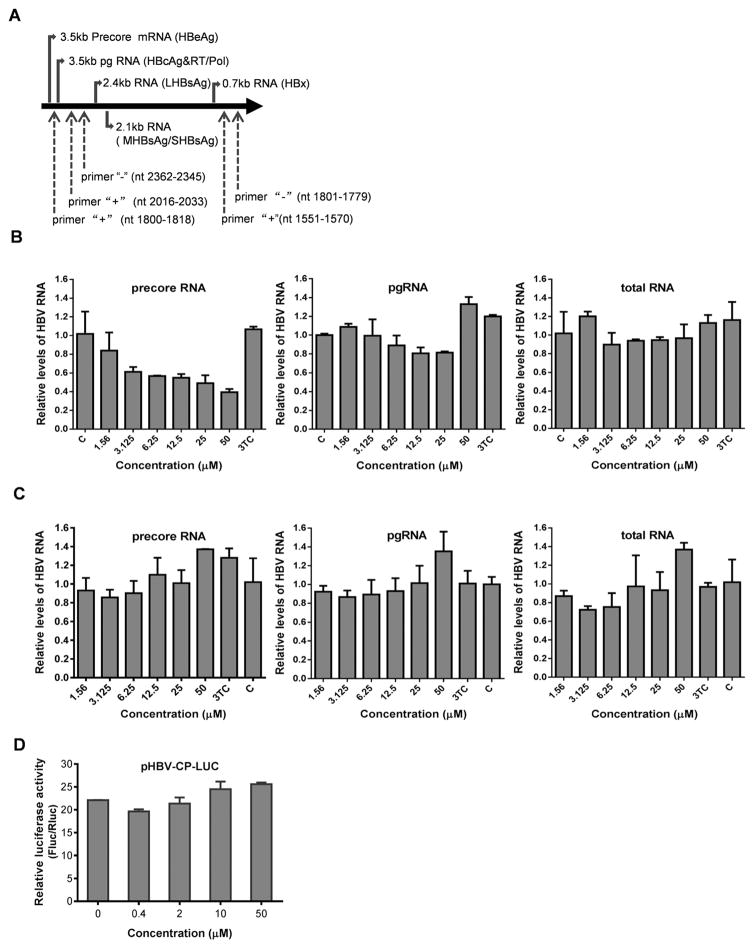
Fig. 3. Punicalagin downregulates levels of precore mRNA in HepG2.117 cells, but has no effect on core promoter activity.
(A) Schematic illustration of HBV RNAs and the PCR primer positions. The primers for precore mRNA detection are primer “+” (nt 1800–1818) and primer “−” (nt 2362–2345). Primer “+” (nt 2016–2033) and primer “−” (nt 2362–2345) were used for pgRNA and precore mRNA amplification. A pair of primers flanking the X ORF region, primer “+” (nt 1551–1570) and primer “−” (nt 1801–1779), were used to amplify HBV total RNA. (B) After induction of HBV replication for three days in the absence of Dox, HepG2.117 cells were treated with various concentrations of punicalagin and lamividine (3TC, 1 μM) for an additional three days and harvested for total RNA extraction. Viral RNAs were detected by real-time PCR. GAPDH was analyzed as an internal reference for normalization purpose. C, untreated control. (C) HepG2.2.15 cells were treated with drugs for 3 days and harvested for total RNA extraction. Viral RNAs were detected by real-time PCR. (D) Twenty four hours after cotransfection with pHBVCP-Luc and pRL-TK, HepG2 cells were treated with different concentrations of punicalagin for 3 days and lysed for dual luciferase reporter analysis.