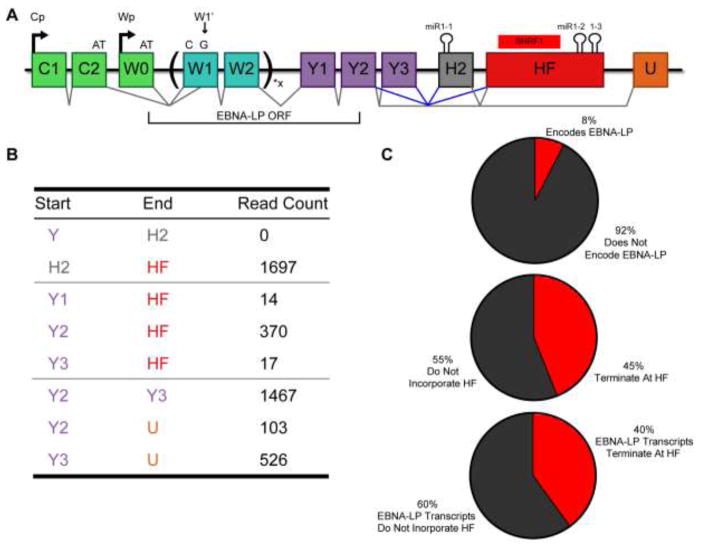
Figure 4.
EBNA-LP encoding transcripts are alternatively spliced to include the HF exon at a high frequency. (A) Schematic of EBNA-LP alternative splicing. EBNA-LP mRNA transcription is driven by either the Cp or Wp promoter. Both promoters encode an exon that terminates with AT (C2 or W0) that must splice to a 3′ splice site located within the first W1 exon, called W1′, to acquire the G to make the ATG necessary to initiate EBNA-LP translation. EBNA-LP transcripts then splice to W2 and can include anywhere from 1 to 11 copies of the W1/W2 repeats. The coding region for EBNA-LP then terminates within the Y2 exon. The H2 exon contains the BHRF1-1 pri-miRNA and the HF exon contains the BHRF1 ORF and pri-miR-BHRF1-2 and 1-3. Known splice junctions are shown in black, including to the U exon that splices 3′ to exons encoding EBNA1, 3A, 3B and 3C. Possible splice junctions that would lead to the inclusion of the miR-BHRF1 miRNAs in the 3′UTR of EBNA-LP are shown in blue. (B) RNA-Seq data from the 1000 genomes project was processed to identify splice junctions. Raw read counts are shown for H2 and HF splicing. Canonical splice junctions are provided at the bottom for reference. (C) SMRT sequencing was performed on RNA isolated using a probe complementary to the W2 exon. Of the 64 complete Cp and Wp driven transcripts sequenced, both the total percentage, and the percentage of the mRNAs encoding EBNA-LP, that terminate at the poly (A) site located at the end of the HF exon,, are shown.