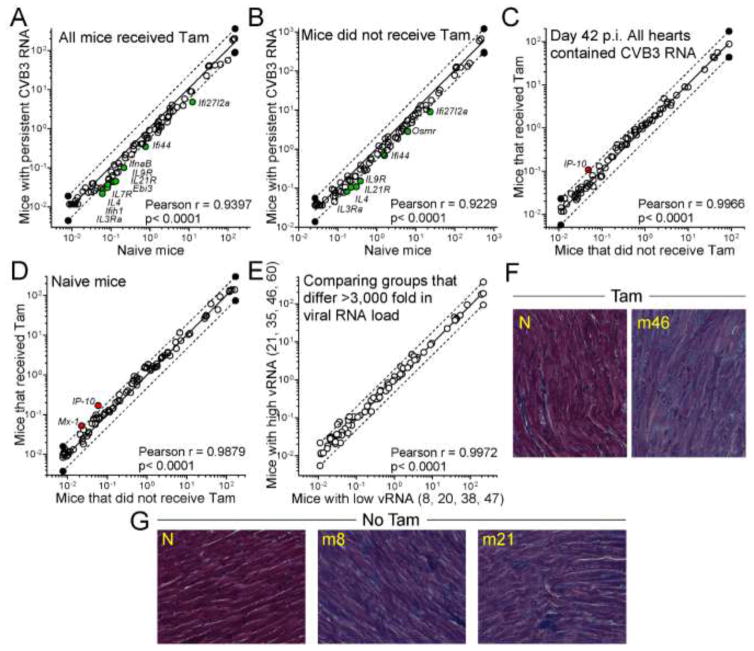
Figure 4. Neither the persistence of CVB3 RNA nor the disruption of T1IFNR expression has a marked impact on IFN-related mRNA abundance or cardiac inflammation.
A – E. PCR array was used to quantify the expression of interferon-related mRNAs in the cardiac RNAs from the selected mice, and from control CMMCM T1IFNRf/f mice. For all comparisons, a linear regression analysis was carried out (solid line in each graph) and 2-fold changes are delimited by dashed lines. Each symbol indicates the relative expression of an individual gene in the two groups shown on the x and y axes. A reduction in mRNA content (y lower than x) is indicated by a green symbol; an increase (y greater than x) is shown by a red symbol. All such changes are annotated with the gene name. None of the changes were flagged as statistically-significant. A. Viral RNA+ Tam-treated mice (y axis) were compared to control Tam-treated mice (x axis), to determine if persistent viral RNA is associated with an interferon signature. B. A similar comparison, this time of mice that did not receive Tam. C. Using the 8 selected RNA+ mice, gene expression in Tam-receiving animals (in which T1IFN signaling is disrupted in cardiomyocytes; y axis) was compared to that in untreated animals (x axis). D. The impact of Tam on gene expression was determined in uninfected mice. E. The effect of viral RNA load was determined by comparing four mice with high load (mean 6.6 × 108 genome copies/g heart, y axis) with four mice with low RNA load (mean 2.1 × 105 genome copies/g heart, x axis) F, G. Histological sections are shown from the indicated d42 mice, and from naïve (N) mice ± Tam (Masson’s trichrome stain).