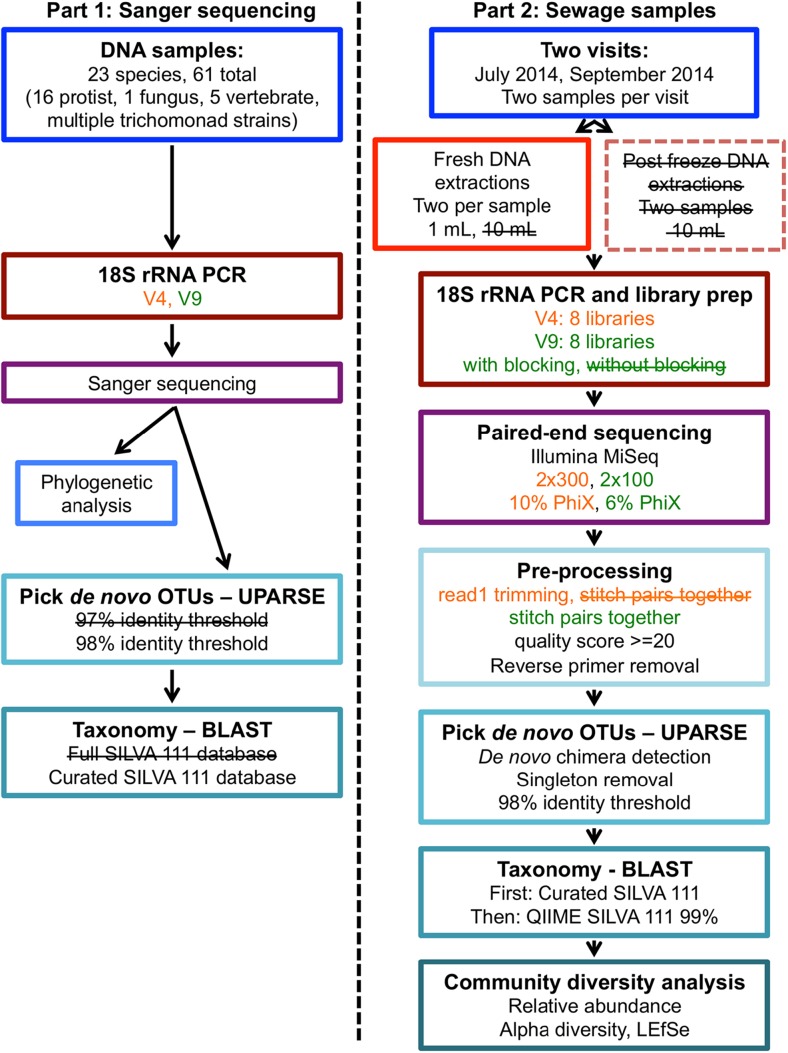
Fig. 1.
Workflow employed for testing and optimization of protocols to detect microbial eukaryotes in raw sewage. Part 1 was used to evaluate two variable regions of the 18S rRNA gene, V4 and V9, and determine best practices for zoonotic protists. Part 2 was used to optimize the methods in Part 1 and develop experimental methods for Illumina sequencing of raw sewage samples. Methods that were tested but determined suboptimal are indicated in boxes with dotted lines and/or crossed out text