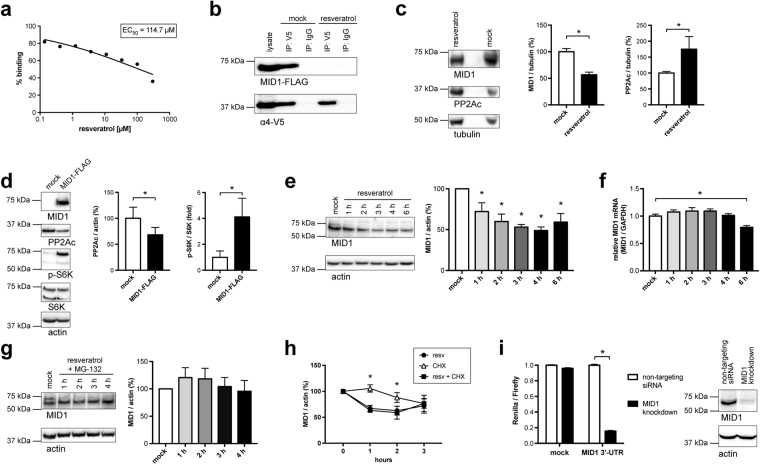
Figure 1.
Resveratrol interferes with the MID1 complex assembly and reduces the MID1 transcript and protein level. (a) AlphaScreen protein-protein interaction assay. Resveratrol in various concentrations (starting at 300 µM) was incubated with MID1 (BBox1/2) and α4 (full-length) coupled to acceptor and donor beads respectively. Upon binding between MID1 and α4 the donor and acceptor beads come into proximity, resulting in a fluorescent signal that was quantified. (b) Co-immunoprecipitation of FLAG-MID1 and α4-V5 using V5 antibodies. Immunoprecipitates were incubated with or without resveratrol and subsequently washed. Immunoprecipitates were analysed on a western blot using FLAG- and V5- antibodies. (c) HEK293-β2a cells were treated with 100 µM resveratrol for 20 hours and analysed on western blots detecting MID1, PP2Ac and tubulin as loading control (n = 3). (d) HEK293T cells were transfected with FLAG-MID1 and analysed on western blots detecting MID1, PP2Ac, phospho-S6K, S6K, and actin as loading control. Right: quantification of western blots. Columns represent mean values +/− SEM (*p < 0.05) (n = 3). (e) HEK293T cells were treated with or without 100 µM resveratrol for 0–6 hours. Left: Cell lysates were analysed on western blots detecting MID1 and actin as loading control. Right: quantification of western blots. Columns represent mean values +/− SEM (*p < 0.05) (n = 3). (f) HEK293T cells were treated with or without 100 µM resveratrol for 0–6 hours. Expression levels of MID1 and GAPDH were analysed by real-time PCR. Samples were measured in triplicates and the relative MID1 expression normalized to GAPDH is shown. Graph represents mean values +/− SEM, (n = 3). (g) HEK293T cells were co-treated with 100 µM resveratrol and 10 µM MG132 for 0–4 hours. Left: Cell lysates were analysed on western blots detecting MID1 and actin as loading control. Right: quantification of western blots. Columns represent mean values +/− SEM (*p < 0.05) (n = 3). (h) HEK293T cells were treated with either the translation inhibitor cycloheximide (50 µg/ml), 100 µM resveratrol, or both substances in combination for increasing time intervals. The MID1 protein levels were analysed on western blots. The graph shows relative MID1 protein levels (normalized to actin), (*p < 0.05). (i) HEK293T cells were co-transfected with either non-silencing control or MID1 specific siRNAs directed against the coding region of MID1 in combination with a plasmid containing the MID1 3′-UTR downstream of the stop codon of renilla luciferase as well as firefly luciferase expressed from a different promoter. Relative light units of renilla normalized to firefly luciferase are shown. Columns represent mean values +/− SEM (*p < 0.01).