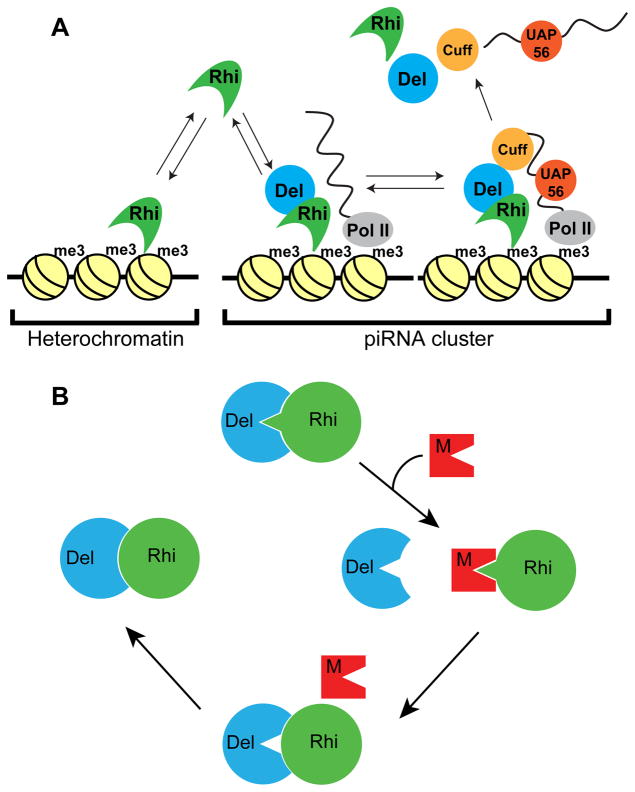
Figure 7. Model for co-evolution of the Rhino-Deadlock interface.
(A) Model for Del function in Rhi localization to piRNA clusters. The Rhi Chromo domain interact with H3K9me3 marks throughout the genome, but most of these marks are in transcriptionally silent regions. At clusters, which are transcribed, Del interactions with Cuff, a putative RNA end binding protein, leads to formation of a chromatin bound complex, which recruits additional RNA binding components (i.e. UAP56). Assembly of these complexes leads to Rhi accumulation at clusters. We further propose that release of piRNA precursor complexes is accompanied by release and recycling of Rhi, Del and Cuff, which then re-initiate the cycle.
(B) A transposon mutation generates a protein that mimics the Del surface that binds to Rhi. Competition for productive Rhi-Del complex formation disrupts piRNA biogenesis and causes increased transposition. Reduced fertility leads to selection of Rhi mutations that reduce mimic binding, at the expense of Rhi-Del affinity. “Leaky” transposition leads to selection of Del mutations that completely restore Rhi binding. Pathogen mimicry thus leads to rapid evolution of Rhi-Del interface.
See also Figure S6.