Summary
Urinary tract infections (UTI) caused by uropathogenic E. coli (UPEC) affect 150 million people annually1,2. Despite effective antibiotic therapy, 30–50% of patients experience recurrent UTI (rUTI)1. Additionally, the growing prevelance of UPEC resistant to last-line antibiotic treatments, and more recently carbapenems and colistin, make UTIs a prime example of the antibiotic-resistance crisis and emphasize the need for new approaches to treat and prevent bacterial infections3–5. UPEC strains establish reservoirs in the gut from which they are shed in the feces, can colonize the peri-urethral area or vagina and subsequently ascend through the urethra to the urinary tract, where they cause UTI6. UPEC isolates encode up to 16 distinct chaperone-usher pathway (CUP) pili and each pilus type likely enables colonization of a habitat in the host or environment7. For example, the type 1 pilus adhesin, FimH, binds mannose on the bladder surface, mediating bladder colonization. However, little is known regarding the mechanisms underlying UPEC persistence in the gut5. Using a mouse model, we found that F17-like and type 1 pili promote intestinal colonization and show distinct binding to epithelial cells distributed along colonic crypts. Phylogenomic and structural analyses reveal that F17-like pili are closely related to pilus types carried by intestinal pathogens, but are restricted to extra-intestinal pathogenic E. coli. Moreover, we show that targeting FimH with a high-affinity inhibitor, mannoside M4284, reduces intestinal colonization of genetically diverse UPEC isolates, while simultaneously treating UTI, without significantly disrupting the the structural configuration of the gut microbiota. By selectively depleting the intestinal UPEC reservoir, mannosides could significantly reduce the rate of UTI and rUTI.
Main Text
The genome of UTI89, a human cystitis isolate, contains 9 distinct functional CUP pili. To determine whether any of these CUP pili promote intestinal colonization, we used a streptomycin mouse model of UPEC intestinal colonization8 to co-colonize C3H/HeN mice with wildtype (WT) UTI89 and one of 9 mutant strains, each lacking a single CUP operon (Extended Data Fig. 1). Deletion of 7 operons yfc, yeh, yad, pap, sfa, yqi, and mat had no effect on UTI89 intestinal fitness compared to the isogenic WT strain (Fig. 1a–g). However, deletion of the fim or ucl pilus operons, which encode type 1 and F17-like pili, respectively, produced significant colonization defects (up to 100- and 1000-fold, respectively; Fig. 1h,i). Loss of FimH, the type 1 pilus adhesin, mirrored the defect caused by deletion of the full type 1 pilus operon (Extended Data Fig. 2a). Deletion of both pilus types in a single strain produced a fitness defect greater than either individual deletion alone, suggesting that these two pilus types do not have redundant roles (Fig. 1j,k).
Figure 1. Type 1 and F17-like pili promote UPEC intestinal colonization.
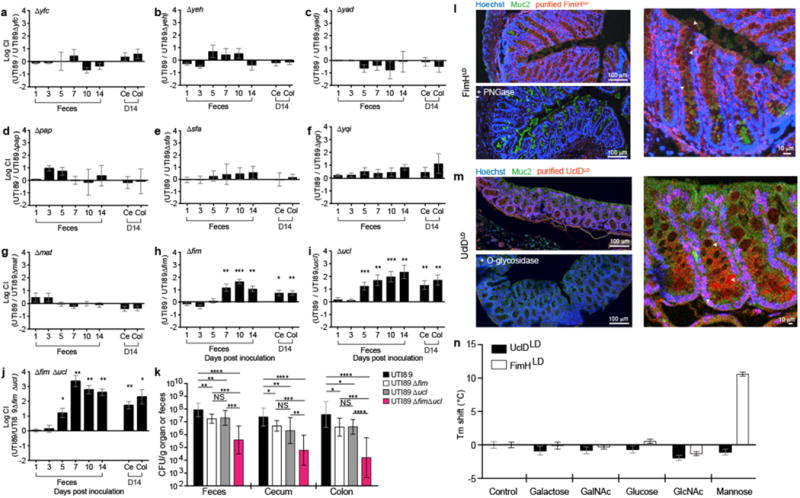
Streptomycin pretreated C3H/HeN mice were concurrently (a–j) or singly (k) colonized with WT UTI89 and/or UTI89 lacking one or more CUP operons. (l,m) Purified adhesin lectin domains FimHLD (type 1 pili) and UclDLD (F17-like pili) were tested for binding to mouse colonic sections. Sections were stained with hoechst (blue) and antibodies to Muc2, a mucus-associated glycoprotein (green). FimHLD and UclDLD binding was lost by pre-treating tissue sections with PNGase and O-glycosidase, respectively. Arrowheads highlight binding by FimHLD or UclDLD. (n) UclDLD does not bind five common monosaccharides. Ce=cecum, Col=colon, CI=competitive index. Bars represent mean values ± SEM (a–j, n), geometric means ± SD (k). *p<0.05, **p<0.01, ***p<0.001 by Wilcoxon Signed Ranked (a–j) or Mann Whitney U test (k,n). n=5 mice, 1 replicate (a, d–g). n=10 mice, 2 replicates (b,h). n=6 mice, 1 replicate (c). n=14 mice, 3 replicates (i). n=8, 2 replicates (j). n=12 mice, 3 replicates (UTI89); n=9 mice, 2 replicates (UTI89Δfim); n=15 mice, 3 replicates (UTI89Δucl); n=10 mice, 2 replicates (UTI89ΔfimΔucl) (k). Replicates are biological (a–k). n=3 tissue sections, 4 represenative images per section (l,m). n=3 wells, 3 technical replicates (n).
In a mouse model, type 1 pilus mediated binding to mannosylated receptors is indispensible for bladder colonization and invasion of urothelial cells lining the bladder lumen2,5. Once inside urothelial cells, a single bacterium rapidly divides, forming an intracellular bacterial community (IBC)2,5. Further, UPEC can access underlying transitional cells, forming quiescent intracellular reservoirs (QIRs)2,5. Mutations in fimH abolish the ability of UPEC to colonize the bladder, form IBCs and QIRs2,5,9. In contrast, no role was observed for F17-like pili in the rate or severity of bladder infection in individual or concurrent transurethral inoculations of UTI89 and UTI89Δucl strains into the bladders of female C3H/HeN mice (Extended Data Fig. 3). Differences between mouse and human bladders or the over-expression of F17-like pili in vitro may account for the variance with another study that showed a role for F17-like pili in binding to desquamated epithelial cells harvested from human urine10.
The fim and ucl operons encode two-domain tip adhesin proteins, FimH and UclD, respectively. The adhesin lectin domain contains the ligand binding site, while the pilin domain joins the adhesin to the pilus rod5. Purified FimH lectin domain (FimHLD) bound to more differentiated epithelial cells located in the upper portion of crypts and in ‘surface epithelial cuffs’ (the colonic homologs of small intestinal villi) (Fig. 1l). FimH binding was prevented by pretreating tissue sections with PNGase, which cleaves N-linked oligosaccharides. FimHLD also bound to Caco-2 cells (an immortalized human enterocyte-like cell line derived from colorectal carcinoma); binding was inhibited by D-mannose and a high affinity mannose-analog (mannoside), M428411 (Extended Data Fig. 2b). The UclD lectin domain, UclDLD, also bound colonic epithelial cells in tissue sections; binding was inhibited by pretreating tissue sections with O-glycosidase, an enzyme that cleaves O-linked oligosaccharides, suggesting that the UclD ligand is contained within an O-glycan (Fig. 1m).
CUP pili are highly conserved throughout Proteobacteria and are assembled by dedicated chaperone-usher assembly machines encoded by each respective CUP operon along with the various subunit types comprising the pilus fiber5,7. The sequence identity between usher genes of distinct CUP pilus types is greater than the identity of genes that encode other CUP pilus proteins and thus can be compared to elucidate evolutionary relationships of CUP pili among Proteobacteria7,12. A homology search of a database of γ-Proteobacteria genomes revealed that the UTI89 F17-like usher gene sequence (uclC) shared highest identity with other E. coli uclC sequences and with orthologous usher sequences of P. mirabilis, a bacterium that can colonize the gut, and S. enterica, an intestinal pathogen (Supplementary Table 1). The uclC usher gene was also closely related to usher genes in F17 (thus the derivation of the name, F17-like), pVir99 and ECs1278 pili. A phylogenetic analysis showed clustering of these E. coli and Proteus species’ ushers into a distinct sub-branch within the broader F17 group usher phylogeny, suggesting that they share a common ancestor (Extended Data Fig. 4). F17-like pili are present in only 10% of E. coli strains; these strains are almost exclusively in the B2 clade, which harbors the majority of extraintestinal pathogenic E. coli (ExPEC) and UPEC strains10,13. In contrast, F17 and ECs1278 pili are found in the intestinal pathogens, enterotoxigenic E. coli (ETEC) and enterohemorrhagic E. coli (EHEC), respectively, which are specific to clades A, E, and B114,15. These findings suggest that UPEC strains in clade B2 may have acquired the ucl operon from a different species and retained this factor to facilitate its residency in the gut. No binding of UclDLD was observed to GlcNAc, the ligand bound by F17 pili, indicating that F17-like pili bind a ligand distinct from that of F17 pili (Fig. 1n)16. Interestingly, although the amino acid sequence of the full length UclD adhesin has diverged from that of the F17 adhesin, F17G, it is virtually invariant across all strains encoding it (>99%), suggesting that there is a single, distinct ligand for UclD adhesins10.
A comparative genomic analysis of 43 strains isolated from a cohort of 14 women at the time of initial presentation with acute UTI, or during subsequent recurrent UTIs revealed that fourteen of the rUTI events were caused by B2 strains (Supplementary Table 2)17,18. Of these 14 strains, 13 encoded F17-like pili (~93%) (Extended Data Fig. 5). In contrast, F17-like pili have been found in less than 50% of all B2 strains10 (E. coli reference collection (ECOR)), suggesting that F17-like pili might be associated with UPEC persistence in women with rUTI due to their ability to promote maintenance of a UPEC intestinal reservoir.
To further characterize F17-like pili, we solved two X-ray crystal structures of UclDLD. The structures in the P21 (green) and P212121 (gray) space groups were resolved to 1.05 Å and 1.6 Å resolution, respectively, and are nearly identical (Fig. 2a-1; Supplementary Table 3). Despite low primary sequence identity (~25%), the structural characteristics of UclDLD and the F17 adhesin, F17GLD are conserved (Fig. 2a–c)16. This includes the presence of a transverse putative binding site in UclDLD located at a similar position to the GlcNAc binding site on F17G (Fig. 2a–3, b, c)16. Structural and sequence alignments reveal two large conserved insertions in UclD relative to F17G (Fig. 2c), whose direct proximity to the putative binding pocket suggests that they are involved in UclD receptor binding. The six conserved residues that make up the candidate binding pocket in UclD are chemically distinct to their F17G counterparts (Fig. 2b,c), providing further evidence that UclD binds a distinct ligand.
Figure 2. Structural analysis of UclDLD.
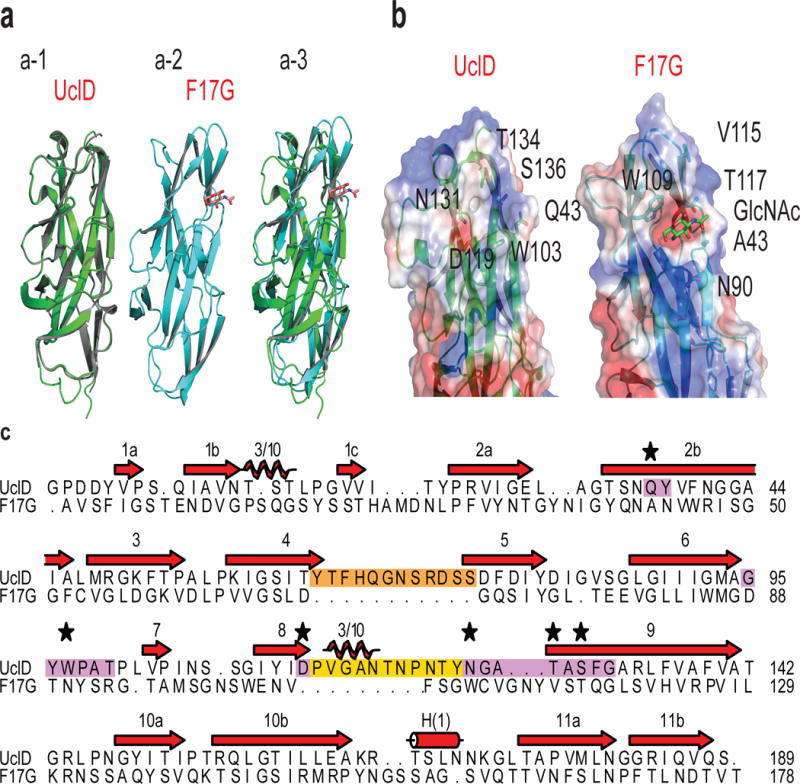
(a) Superposition of the P21 UclDLD (green) and P212121 UclDLD (grey) crystal structures (a-1). F17G adhesin crystal structure (PDB 1OIO16) (a-2). Superposition of P21 UclDLD (green) and F17GLD (cyan) structures (a-3). (b) Comparison of residue positioning and electrostatic surface potential of the putative binding site between the UclDLD structures and the known binding site of F17GLD. (c) Structural alignment of UclDLD and F17GLD amino acid sequences. Purple residues highlight the putative UclDLD binding site. Orange and yellow residues highlight insertions in UclDLD sequence. Starred residues are hypothesized to mediate UclD ligand binding. Beta strands (red arrows), 3/10 helices (coils), and alpa-helices (cylinder) are shown.
Figure 3. Mannoside simultaneously reduces the UPEC intestinal reservoir and treats UTI.
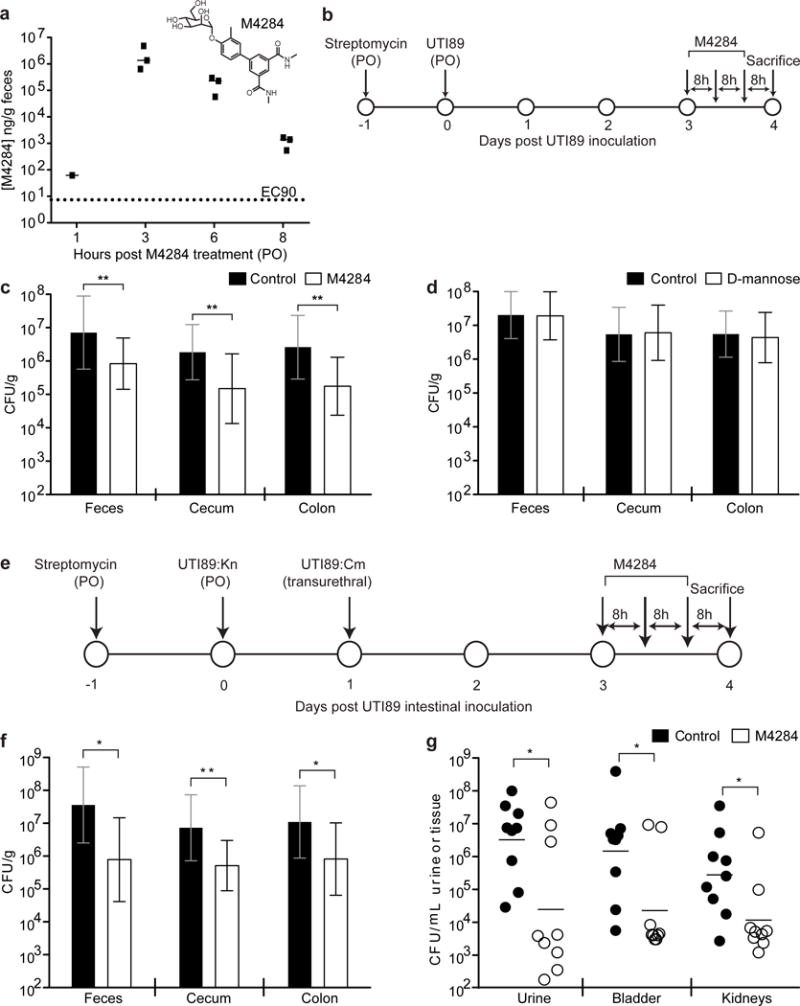
(a) M4284 concentration in mouse feces after 1 dose (100mg/kg administered by oral gavage). (b) C3H/HeN mice were intestinally colonized with UTI89 and given 3 oral doses of M4284 (100mg/kg), vehicle alone (10% cyclodextrin, Control), or D-mannose (100mg/kg). (c,d) UTI89 levels in the feces and intestinal segments. (e) UTI89 was introduced into the gut of C3H/HeN mice by oral gavage and into the bladder by transurethral inoculation before receiving 3 doses of M4284. (f,g) UTI89 levels in the gut and urinary tract were assessed. Bars represent median (a), geometric means ± SD (c,d,f); geometric mean (g) *p<0.05, **p<0.01 by Mann Whitney U test. n= 3 mice, 1 replicate (a). n=14 mice (Control); n=15 mice (M4284); 3 replicates (c). n= 10 mice, 2 replicates (d). n=9 mice, 2 replicates (f, g). All replicates biological (a–g).
In light of the role of FimH and UclD in gut colonization, we conducted a study designed to reduce the UPEC intestinal reservoir with an adhesin-directed therapeutic. M4284 is a high-affinity biphenyl mannoside whose binding affinity for FimH is ~100,000-fold higher than the natural sugar, D-mannose11,19. Pharmacokinetic (PK) analysis revealed that M4284 concentrations remain high in the feces of mice for up to 8 h after an oral dose (Fig. 3a). Treating mice that were colonized by UTI89 with 3 doses (administered by oral gavage) of M4284, a regimen that successfully treats UPEC UTI in mice20, significantly reduced UTI89 levels in feces, cecum, and colon compared to those treated with a vehicle control (Fig. 3b,c). Treating mice with additonal M4284 doses further reduced the UTI89 population and the overall number of UPEC continued to be lower in M4284-treated mice after termination of treatment (Extended Data Fig. 6). While D-mannose blocks FimH binding in vitro, treating mice with D-mannose did not alter UTI89 levels in vivo (Exended Data Fig. 2b, Fig. 3d).
The ID50 in the UTI mouse model is 105 CFU21. Furthermore, decreasing the dose of UPEC introduced into the bladder from 108 to 106 CFU significantly reduced the rate of UTI, suggesting that the 1–1.5 log (or 90–95%) mannoside-driven reduction in fecal UPEC levels would reduce the numbers of bacteria available to access the urinary tract and likely reduce the rate of UTI and/or rUTI (Extended Data Fig. 7). Indeed, we found that M4284 simultaneously reduces UTI89 levels in the gut and urinary tracts of mice that were concurrently colonized with UTI89 in the gut and bladder (Fig. 3e–g).
We sequenced bacterial 16S rRNA gene amplicons generated from fecal samples of C3H/HeN mice that had not been given streptomycin or infected with UPEC but were treated with 3 doses of M4284 or vehicle alone. We found that M4284 produced no significant changes in the overall phylogenetic configuration of the microbiota as judged by the unweighted UniFrac dissimilarity metric, in contrast to the significant perturbations produced by treatment with ciprofloxacin, a fluoroquinolone antibiotic (Fig. 4a, Extended Data Fig. 8a). Using this UniFrac metric, we found that M4284 treatment did not produce significant perturbations in bacterial community structure in mice pretreated with streptomycin and then colonized with UTI89 (Extended Data Fig. 9). We concluded that M4284 can function to selectively extirpate UPEC from the gut in our preclinical model. Interestingly, while most Enterobacteriaceae carry the fim operon7,18,22, M4284 treatment does not significantly affect the abundance of intestinal Enterobacteriaceae (Extended Data Figure 8a,b), suggesting that these bacteria may not be expressing type 1 pili during M4284 exposure or that they reside within inaccessible intestinal habitats.
Figure 4. Mannoside treatment minimally effects the fecal microbiota configuration and targets human UPEC isolates in mice with different genetic backgrounds.
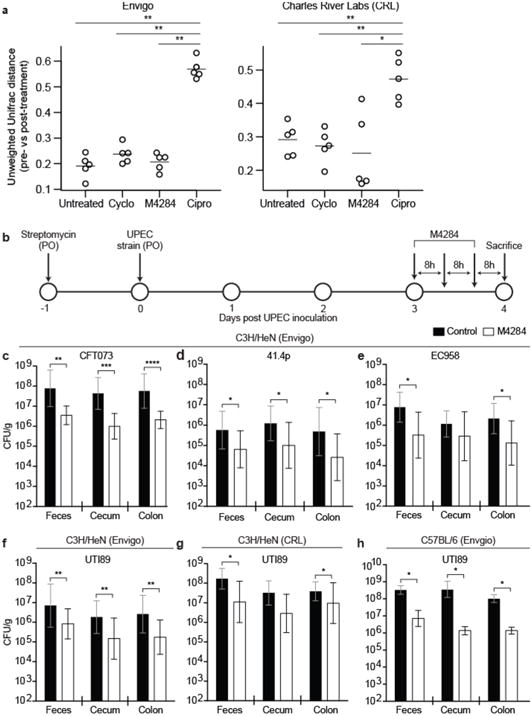
Mice were given one of the following oral treatments: (i) M4284 (100mg/kg), (ii) cyclodextrin (Cyclo, 10%), (iii) ciprofloxacin (Cipro, 15mg/kg), or (iv) none (Untreated). Fecal community structure was defined by sequencing bacterial 16S rRNA gene amplicons. (a) For each treatment performed on C3H/HeN mice from Envigo or CRL, the change in microbiota configuration was determined by measuring the unweighted UniFrac distance between samples obtained from each animal before treatment and 24 h after the last dose (larger UniFrac distance equates to a larger shift in community structure). (b–f) Mice were colonized by oral gavage (PO) of one of four different UPEC strains and given three doses of M4284. (g,h) The ability of M4284 to target UTI89 in C3H/HeN mice from CRL (g) and C57BL/6 mice from Envigo Labs (h) was also assessed. Bars represent median (a) or geometric means ± SD (c–h). *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 Mann Whitney U test. n=5 mice per vendor, 1 replicate (a). n=10 mice, 2 replicates (c–e, g). n=14 (Control, 10% cyclodextrin); n=15 (M4284); 3 replicates (f). n=10 mice (Control, 10% cyclodextrin); n=9 mice (M4284); 2 replicates (h). All replicates are biological (a–h).
M4284 treatment of mice colonized with three additional genetically diverse UPEC clinical isolates (EC95823, 41.4p18 and CFT07324), reduced the levels of each UPEC strain by a similar percentage in the feces, cecum, and colon (Fig. 4b–f, Extended Data Fig. 10a). Furthermore, we found that M4284 treatment reduced UTI89 levels in C3H/HeN and C57BL/6 mice from different vendors, harboring distinct gut microbial communities. In each case tested, the percentage reduction in UPEC levels in cecum, colon and feces did not vary significantly between the different treatment groups (Fig. 4f–h, Extended Data Figs. 8c,10b). We concluded that M4284 treatment has activity against different UPEC strains in different host genetic backgrounds and gut microbial community contexts.
Prospectus
As the prevalence of antibiotic-resistant pathogens continues to rise, the need to develop highly targeted/specific therapeutic approaches has gained increased urgency25,26. Additionally, an increasing number of studies are finding that disruption of the gut microbiota by orally administered antibiotics, especially during childhood, may affect its functional properties in ways that are deleterious to the host, not only in the short term but for more protracted periods of time27,28. Therefore, developing therapeutic agents, like mannosides, that specifically target a pathogen without disrupting the remainder of a microbial community has important ramifications not only for UPEC but potentially for other infections, including those caused by enteropathogens. In addition, identification of genes involved in UPEC intestinal colonization may also provide a method by which UTI patients could be stratified for epidemiologic studies of risk for recurrent disease as well as for proof-of-concept clinical studies of the efficacy of CUP-directed treatment regimens.
Materials and Methods
Ethics statement
The Washington University Animal Studies Committee approved all procedures used for the mouse experiments described in the present study. Overall care of the animals was consistent with The Guide for the Care and Use of Laboratory Animals from the National Research Council and the USDA Animal Care Resource Guide. For collection of colonic tissues for adhesion binding studies, mice were sacrificed according to institutional, national and European animal regulations, using protocols that were also approved by the animal ethics committee of Ghent University.
Bacterial strains
CUP operon and adhesin deletions in UTI89 were engineered by replacing the gene(s) of interest with antibiotic-resistance markers using the λ Red Recombinase system30. Earlier reports described WT UTI89 and its isogenic fim and fimH mutants21,31 as well as EC95823, 41.4p18 and CFT07324.
Colonization of mice with UPEC strains
6-week old female C3H/HeN mice were obtained from Envigo or Charles Rivers Labs (CRL). 6-week old female C57BL/6 mice were also obtained from Envigo. Animals were maintained in a single room in our vivarium for no more than 2 days prior to treatment. Prior to and after treatment all animals received PicoLab Rodent Diet 20 (Purina) ad libitum. All animals were maintained under a strict light cycle (lights on at 0600h, off at 1800h). For competitive infections, if a phenotype was observed after testing five mice (1 biological replicate), the experiment was repeated 1–2 times (total of n=10–16 mice, 2–3 biological replicates). For 16S rRNA analyses, 4–5 mice were examined (1 biological replicate). For all other experiments, 9–16 mice were tested and the experiment was repeated 2–3 times (2–3 biological replicates). Exclusion criteria for mice were pre-established; (i) both introduced strains in competitive infections became undetectable during the course of a 14 day experiment, and (ii) mice died or lost > 20% of their body weight. No mice in this study met these criteria. Mice were acquired from indicated vendors and randomly placed into cages (n=5 mice/cage) by employees of Washington University’s Division of Comparative Medicine (DCM); no additional methods for randomization were used to determine how animals were allocated to experimental groups. Investigators were not blinded to group allocation during experiments.
Animals received a single dose of streptomycin (1000mg/kg in 100 μL water by oral gavage (PO)) followed 24 h later by an oral gavage of ∼108 CFU UPEC in 100 μl phosphate-buffered saline (PBS). Bladder infections were performed via transurethral inoculation32. UPEC strains were prepared for inoculation as described previously32. Briefly, a single UTI89 colony was inoculated in 20 mL of Luria Broth (LB) and incubated at 37°C under static conditions for 24 h. Bacteria were then diluted (1:1000) into fresh LB and incubated at 37°C under static conditions for 18–24 h. Bacteria were subsequently washed three times with PBS and then concentrated to ~1×108 CFU per 100 μL for intestinal infections and ~1×108 CFU per 50 μL for bladder infections.
In all cases, fecal and urine samples were collected directly from each animal at the indicated time points. Fecal samples were immediately weighed and homogenized in 1 mL PBS. Urine samples were immediately diluted 1:10 prior to plating. Mice were sacrificed via cervical dislocation under isofluorane anesthesia and their organs were removed and processed under aseptic conditions. Intestinal segments (cecum and colon) were weighed prior to homogenization and plating on LB supplemented with the appropriate antibiotic.
Enumeration of bladder intracellular bacterial communities (IBCs)
6 week old C3H/HeN mice were given a single oral dose of either M4284 (100mg/kg) or vehicle control (10% cyclodextrin) 30 min before transurethral inoculation with UTI89. To accurately count the number of IBCs, mice were sacrified 6 hours after infection. Bladders were removed aseptically, bi-sected, splayed on silicone plates and fixed in 4% (v/v) paraformaldehyde. IBCs, readily discernable as punctate violet spots, were quantified by LacZ staining of bladder wholemounts20,33.
Immunofluorescence Studies
The protocols used for immunohistochemical analysis are based on a previous study34. Following sacrifice of 6 week old, female C57BL/6 mice (supplied by VIB-Ghent University breeding program, Belgium), segments of colon were fixed in methanol-Carnoy for a minimum of 3 hours at room temperature. The fixed tissues were then embedded in paraffin and 4-μm-thick sections were cut and placed on glass slides. Slides were de-paraffinized and re-hydrated by incubating them in xylene, isopropanol, 100% ethanol and finally 70% ethanol (each step involving a 3 min incubation in the reagent followed by another 3 min incubation in fresh reagent). Slides were subsequently rinsed in tap water and PBS, placed in blocking buffer (5% fecal calf serum prepared in PBS) at room temperature for 30 min, and then incubated with rabbit polyclonal antibodies to Muc2 [1/2000; Mucin 2 (H-300), Santa Cruz Biotechnology] for 2 h. After three washes with PBS, slides were incubated with a goat anti-rabbit Dylight-488 labeled secondary antibody (1/1000 dilution, ThermoFisher Cat No. 35553) in blocking buffer for 1 h at room temperature. Slides were washed three times with PBS before counterstaining with bis-benzimide (Hoechst dye) (1/1000 in PBS) for 10 min at room temperature. Finally, slides were incubated with FimHLD or UclDLD (P21) protein, labeled with NHS 650nm Dylight, in blocking buffer at 4°C overnight. Prior to staining, sections were treated with O-glycosidase (NEB) or PNGaseF (SIGMA) at 37°C using buffers and protocols supplied by manufacturer. Slides were washed subsequently with PBS before treatment with fluoro mounting medium (n-propyl gallate in glycerol) and viewing under a confocal microscope (Leica Microsystems LAS-AF-TCS SP5) using a 20×125 objective.
Mannoside Treatment
D-mannose or the mannoside M4284 (which has been characterized in a prior published study11), were diluted in vehicle (water and 10% cyclodextrin, respectively) and administered to 6 week old C3H/HeN mice at a dose of 100mg/kg. Control animals were treated with water or 10% cyclodextrin alone. Unless stated otherwise, three doses of M4284, cyclodextrin, or D-mannose were given via oral gavage over 24 hours, with doses administered 8 h apart. Mice were sacrificed and intestinal tissues were processed for analysis of viable bacteria (CFU) 8 h after the last dose, unless otherwise noted. To test the effect of M4284 on intestinal UPEC titers after treatment was terminated, mice were sacrificed 5 days after the last dose of mannoside. To test the effect of additional doses on M4284 treatment on UPEC titers, mice were given 5 doses of mannoside; the first 3 doses were administered 8 h apart, followed 12h later by the 4th dose, and 24h later by the 5th dose. Mice were sacrificed 24h after the 5th dose.
Carriage of F17-like pili
We examined 43 available UPEC isolates (Supplementary table 2). These isolates originated from a clinical study of 14 women who experienced at least two episodes of UTI (an initial UTI and one or more rUTI) during the 90 day study window17. The isolates used in this work were sequenced in a previous study18 (Bioproject ID: PRJNA269984) and include (i) 14 isolates collected at enrollment, (ii) 18 isolates collected during rUTI (10 women experienced a single rUTI while four women experienced two rUTI events), and (iii) 11 isolates collected in the days leading up to an rUTI.
The distribution of the F17-like operon in these clinical E. coli isolates was determined using BLAST and the F17-like operon from UTI89 as the query sequence. A “hit” was considered as any genome sequence that matched the entire length of the query sequence with >75% identity. As a control to prevent false negatives in the BLAST search of draft genomes, DNA sequencing reads from each clinical UPEC isolate were mapped against a reference sequence constructed by concatenating all the ucl genes with 100 N’s separators using Geneious v6.1.735.
Phylogenetic Analyses and Sequence Alignments
Amino acid alignments of full-length UTI89 UclD, P. mirabilis UcaD, S. enterica UclH, and ETEC F17G were conducted using the MAFFT L-INS-i iterative refinement method and the default BLOSUM62 scoring matrix (Supplementary table 1)36. MAFFT collected up to 100 homologs with E values of less than 1e-10 to each sequence from the SwissProt database to improve alignment accuracy. Homologs are automatically removed from the final alignment. The alignment was visualized using Geneious35. A homology search of the coding sequence database of the European Nucleotide Archive (ENA) was conducted using the Basic Local Alignment Search Tool (BLAST)37 using the UTI89 uclC (ENA accession: ABE10308) and EDL933 ECs1278 (ENA accession: AIG67653) as queries. Sequences that matched either gene sequence with >50% identity were downloaded and then filtered to remove partial hits (<80% length of query sequence) and sequences with nonsense mutations, which resulted in a total of 659 sequences (Supplementary table 1). Duplicate sequences were then removed, resulting in a list of 122 unique, representative sequences. These sequences were then aligned with the UTI89 fimD usher sequence (ENA accession: ABE10417) as an outgroup using the MAFFT MAFFT L-INS-i alignment method and the 200PAM scoring matrix36: The phylogenetic relationship between gene sequences was then estimated using RAxML v8.1.3 with the GTRCAT model and supported with 1000 bootstrap replicates; the tree was visualized using the tool interactive Tree of Life (iTOL) v338.
Enzyme-linked immunosorbent assay (ELISA) targeting FimH
Caco-2 cells (ATCC Number HTB-37) were cultured in minimum essential medium (MEM) supplemented with 20% fetal bovine serum (FBS). Cell cultures tested negative for mycoplasma. Cells were split into 48-well plates, grown to 100% confluence and then fixed with paraformeldahyde for 15 min followed by treatment with blocking buffer (1x PBS containing 2% BSA) for 2h. A truncated FimH, corresponding to residues 1–178 of the mature FimH adhesin (FimHLD), expressed in E. coli and purified as described previously39, was serially diluted in blocking buffer and incubated with the fixed CaCo-2 cells for 1h at room temperature. To test the effect of D-mannose or M4284 on FimH binding, 0.2 mg/mL FimHLD was pre-incubated for 5 min in the presence or absence of 1 mM D-mannose (Sigma-Aldrich) or 1mM M4828 (in 20 mM Tris pH 8.0 or 20 mM Tris plus water or 10% cyclodextrin, respectively) before serial dilution and incubation. Wells were washed four times with PBS/0.05% Tween 20 (PBST) prior to incubation with a polyclonal rabbit anti-T3 antibody against FimHLD (generated against FimH residues 1–165; ref. 39) for one hour at room temperature. Following another series of four washes, secondary antibody (goat anti-rabbit Ig conjugated to horse radish peroxidase; ThermoFisher, Catalog Number 32460) was incubated with the cells for 1 h at room temperature (24°C) before washing in PBST. Plates were developed with the BD OptEIA TMB substrate reagent kit for 5 min at room temperature (24°C) before quenching with 1 M H2SO4. Binding was assessed by measuring the absorbance at 450 nM on a TECAN infinite 2 PRO plate reader. Wells lacking protein were used as controld. All conditions were examined in quadruplicate.
Effect of antibiotic exposure on the microbiota
Six week old female C3H/HeN mice from Envigo and CRL were subjected to the following treatments: (i) none (naïve control mice, Untreated), (ii) three doses of M4284 (100mg/kg; in 10% cyclodextrin) or 10% cyclodextrin given 8 h apart, or (iii) ciprofloxacin (two doses of 15mg/kg given 12 h apart). All doses were given via oral gavage. Five mice were included for each treatment type (1 biological replicate) and fecal samples were collected prior to treatment and 24 hours after the last dose of each treatment. Another group of four C3H/HeN mice from Envigo (1 biological replicate) were pretreated with streptomycin and colonized with UTI89 before recieving treatment with either three doses of M4284 (100mg/kg; in 10% cyclodextrin) or with 10% cyclodextrin alone; fecal samples were collected prior to treatment but after exposure to streptomycin and UTI89 and 24 hours after the last dose of each treatment.
Bacterial 16S rRNA sequencing
DNA was extracted by bead beating in extraction buffer [200 mM Tris (pH 8.0), 200 mM NaCl, 20 mM EDTA], 210μL of 20% SDS and 500μL phenol:chloroform:isoamyl alcohol (pH 7.9, 25:24:1). This crude DNA extract was purified (Qiaquick PCR purification kit) and PCR used to generate amplicons from the V4 region of bacterial 16S rRNA genes using primers and cycling conditions described previously40. Amplicons were pooled in equimolar ratios and sequenced on an Illumina MiSeq instrument (paired end 250 nt reads). Paired V4-16S rRNA sequences were merged using FLASH software41, demultiplexed, and reads clustered into 97%ID OTUs (2013 Greengenes OTU reference database; QIIME version 1.9.042). A custom database using modified NCBI bacterial taxonomy was used to train the Ribosomal Database Project (RDP) version 2.4 classifier and assign taxonomy to picked OTUs43. The resulting OTU table was filtered to include only OTUs found in at least two samples at greater than or equal to 0.1% relative abundance.
F17-Like Constructs and Purification
For the P21 UclDLD construct, the first 197 amino acids of the mature UclD adhesin protein were cloned into pDEST14 using Gateway® technology (Invitrogen), resulting in plasmid pUclDAD. Expression was induced with 1mM IPTG. Periplasmic extracts were prepared by resuspending bacterial pellets in 20mM Tris/20% sucrose (pH 8) (4 ml/per gram of pellet). Subsequently, 40μl of 0.5M EDTA and 10mg/ml lysozyme were added per gram of pellet and the suspension was incubated on ice for 30 min. This step was followed by addition of 40 μl of 2.5M MgCl2 per gram of cell pellet and incubation on ice for 5 min. Cells were spun at 15,000 × g and the supernatant was saved as the periplasmic extract. The extract containing the UclD lectin domain was dialyzed against 20mM HEPES pH 7, passed over a SP FF cation exchange column (GE) and bound material eluted with 20mM HEPES pH7/1M NaCl. Pooled fractions containing UclD lectin domain were then applied to a Phenyl Hi Trap column (GE) after addition of 1M ammonium sulfate. Elution was performed using 20mM HEPES pH 7.
To generate purified UclDLD for the P212121 space group, DNA from the UTI89 uclD gene encoding the N-terminal 217 amino acids of the protein were cloned into pTRC99a with a C-terminal 6-His tag. This construct was expressed in the periplasm of E. coli DL41(DE3), a methionine auxotroph strain suitable for expression of native or selenomethionine-labeled protein. Periplasmic extracts were first dialyzed against PBS supplemented with 250 mM NaCl, then bound to a Cobalt (Goldbio) column; bound proteins were eluted with PBS containing 250 mM NaCl, and 250 mM imidazole. Pooled fractions were dialyzed into 20 mM MES (pH 5.8), bound to an HR16/10 Mono S cation exchange column (GE Healthcare), and eluted with 300 mM NaCl. Following cleavage of the periplasmic localization sequence, the mature form of UclDLD-6×His contained 203 amino acids.
Selenomethionine-labeled protein was purified using the same protocol, but all buffers were supplemented with 2 mM β-mercaptoethanol and 1 mM EDTA to prevent oxidation. EDTA was omitted from the periplasmic dialysis buffer to prevent chelation of immobilized cobalt.
Crystallization and Structure Determination
For the P21 UclDLD structure solved in the P21 space group, UclDLD (15mg/ml) was crystallized using sitting drop vapor diffusion against a solution containing 16% PEG 4000, 0.1M Tris HCl pH 8.5, 0.2M magnesium chloride. UclD crystals were flash cooled to 100°K in a solution containing 16% PEG 4000, 0.1M Tris HCl pH 8.5, 0.2M magnesium chloride and 30% glycerol. Data were collected at beamline ID29 (ESRF, Grenoble, France) to 1.05 Å resolution. Data were indexed and processed with XDS44, scaled and merged using SCALA in the CCP4 suite45. Data and refinement statistics can be found in Supplementary Table S3.
For the UclDLD structure solved in the P212121 space group, UclDLD (10 mg/ml 10 mM MES 5.8) was crystallized by the hanging drop vapor diffusion method against a well solution containing 0.1 M potassium phosphate (monobasic), 0.2 M potassium iodide and 20% PEG 3350. One microliter of the protein solution was mixed with 1 μL well solution and incubated at 18 °C. Crystals were harvested and transferred to a solution containing 0.1 M KH2PO4, 0.2 M KI and 20% PEG 3350 supplemented with 20% glycerol before being flash-frozen in a bath of liquid N2. Data were collected at beamline 4.2.2 (ALS Berkeley) to 1.6 Å resolution. Data were indexed and processed with XDS44, scaled and merged AIMLESS in the CCP4 suite45 and phased with the Single anomalous dispersion (SAD) method using phenix.autosol, and refined with phenix.refine46. Data and refinement statistics can be found in Supplementary Table S3. RMSD values were calculating using the DALI server47. Structural alignments were performed in Promal’s 3D using the default settings. Secondary structure assignments for UclDLD were completed using DSSP.
Differential Scanning Fluorimetry
Purified UclDLD (1.4 μg/well) was incubated with 5× Sypro orange fluorescent dye in 20 mM Tris (pH 8.0) with or without 10 mM monosaccharide in a total volume of 70 μL. Samples were heated from 20 °C to 100 °C in 30-second/0.5 °C increments using a Bio-rad C1000 thermocycler with CFX96 RT-PCR attachment. The reported melting temperatures were determined by the inflection point of the sigmoidal graph.
Data Availability
Bacterial V4-16S rRNA datasets have been deposited in the European Nucleotide Archive (ENA) under accession number PRJEB19121. Sequences used to examine the carriage of F17-like pili in clinical rUTI isolates were previously published18 and are deposited in the NCBI under the Bioproject ID: PRJNA269984. Crystallography data have been deposited in the PDB under accession codes: 5NWP (P21) and 5VQ5 (P212121).
Code Availability
No new code was generated for this study. All software was obtained from publicly available sources; papers describing the software are cited in the text.
Statistical Analysis
The statistical significance of differences between groups in experiments (excluding competitive infections) was determined by a Mann Whitney U test. Competitive Index (CI) was defined as (CFU output strain A/CFU output strain B)/(CFU input strain A/CFU input strain B). For competitive infections, statistical significance was determined by a Wilcoxon Signed Ranked test. Statistical analyses were performed using Graphpad Prism 7.
Extended Data
Extended Data Figure 1. Streptomycin treatment allows for persistent UTI89 colonization of the cecum and colon in female C3H/HeN and C57BL/6 mice.
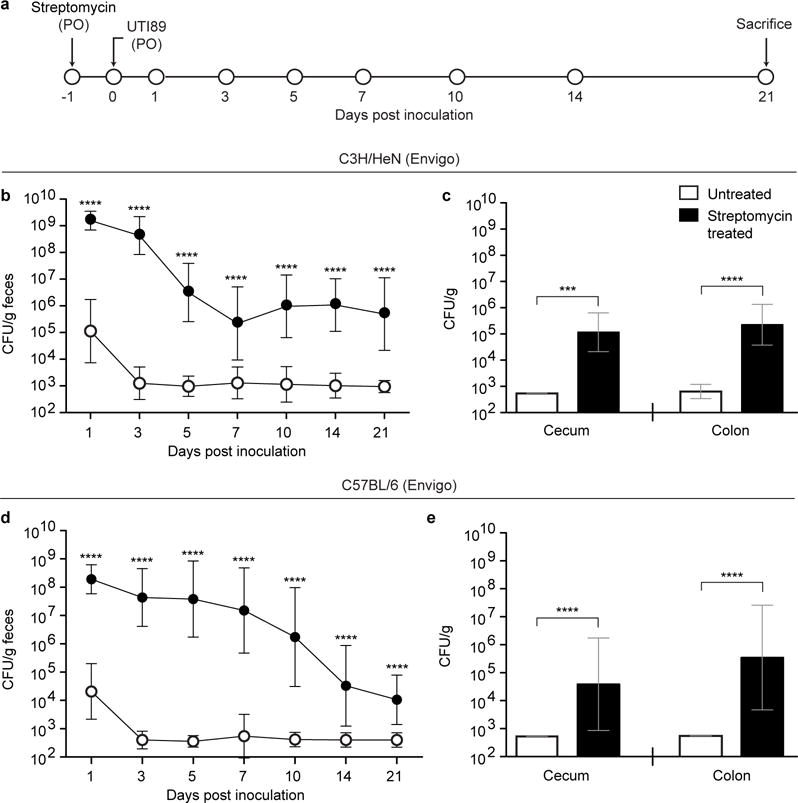
(a) Mice were pretreated with streptomycin and subsequently colonized via oral gavage (PO) with UTI89, a prototypical human UPEC cystitis isolate. (b–e) Colonization of UTI89 in C3H/HeN mice from Envigo (b,c) or C57BL/6 mice from Envigo (d,e) was assessed by quantifying colony forming units (CFU) in fecal samples collected over the course of 21 days from mice who did not receive streptomycin (white circles) or mice pretreated with the antibiotic (black circles). CFU analysis of levels of colonization in the cecum and colon were defined by analyzing tissue homogenates prepared 21 days post colonization. Symbols represent geometric means ± SD, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 (Mann Whitney U test). n=15 mice, 3 biological replicates (b–e).
Extended Data Figure 2. The FimH adhesin is required for type 1 pilus-dependent colonization of the mouse gut and for binding to human intestinal epithelial cells.
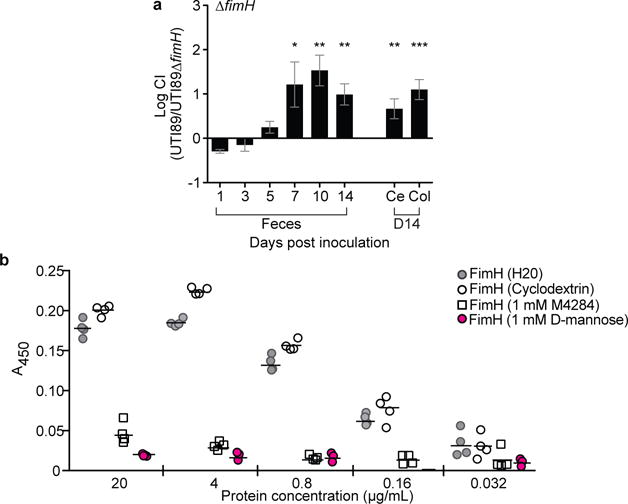
(a) C3H/HeN mice from Envigo were pretreated with streptomycin and concurrently colonized with 1×108 CFU of WT UTI89 and UTI89ΔfimH. The WT strain outcompetes the strain lacking the FimH adhesin. (b) The ability of purified FimH lectin domain (FimHLD) to bind to Caco-2 cells was assessed by a FimH-ELISA. Pre-incubation of FimHLD with D-mannose (1mM) or M4284 (1mM) results in significant reductions in FimH binding to Caco-2 cells while 10% cyclodextrin (M4284 vehicle) had no significant effect. All data shown are normalized to wells that were not exposed to the purified adhesin. Abbreviations CI= competitive index. Ce= cecum, Col= colon. Bars represent mean values ± SEM, *p<0.05, **p<0.01, ***p<0.001, Wilcoxon Signed Ranked test in (a). Bars represent median (b). n=14 mice, 3 biological replicates (a); n=4 wells examing FimH binding to Caco-2 cells per protein concentration, 4 technical replicates (b).
Extended Data Figure 3. F17-like pili are not required for UTI in mice.
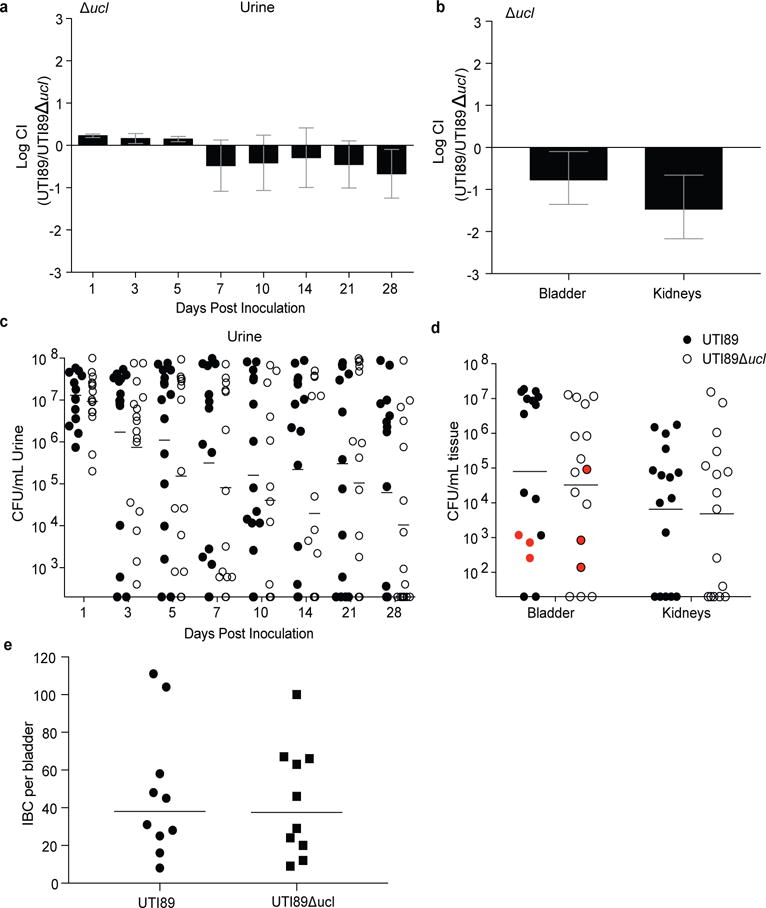
C3H/HeN mice received a transurethral inoculation of UTI89 (WT) and UTI89Δucl, concurrently (a, b), or individually (c–e). (a) UTI89Δucl and WT strains persist at similar levels in the urine over 28 d in competitive infections. (b) The two strains are also present at equal levels in the bladder and kidney at the time of sacrifice (28 days post infection). (c) Single infection with the WT strain (black circles) or the F17-like mutant strain (white circles) produces similar levels of bacteruria over 28 days. (d) Single strain infection also produces similar levels of viable cells in homogenates of whole bladder or kidneys harvested at the time of sacrifice (28 days post infection). There was no statistically significant difference in the number of mice that resolved bacteriuria while maintaining bladder-associated CFUs after transurethral infection with either WT or UTI89Δucl (highlighted in red in d), suggesting that both strains are capable of forming similar numbers of QIRs. (e) Mice infected transurethrally with WT or Δucl strains of UTI89 exhibit a similar number of intracellular bacterial communities (IBCs) at 6 hours in the bladder, indicating that loss of the ucl operon does not alter UTI89’s ability to form IBCs. CI= competitive index. Bars represent mean ± SEM (a,b), geometric mean (c,d) or median (e). No significant difference was detected between any samples by Wilcoxon Signed Ranked test (a,b) or Mann Whitney U test (c–e). n=10 mice, 2 biological replicates (a–b, e). n=16 mice, 3 biological replicates (c, d).
Extended Data Figure 4. Distribution of F17 usher homologs in members of Enterobacteriaceae.
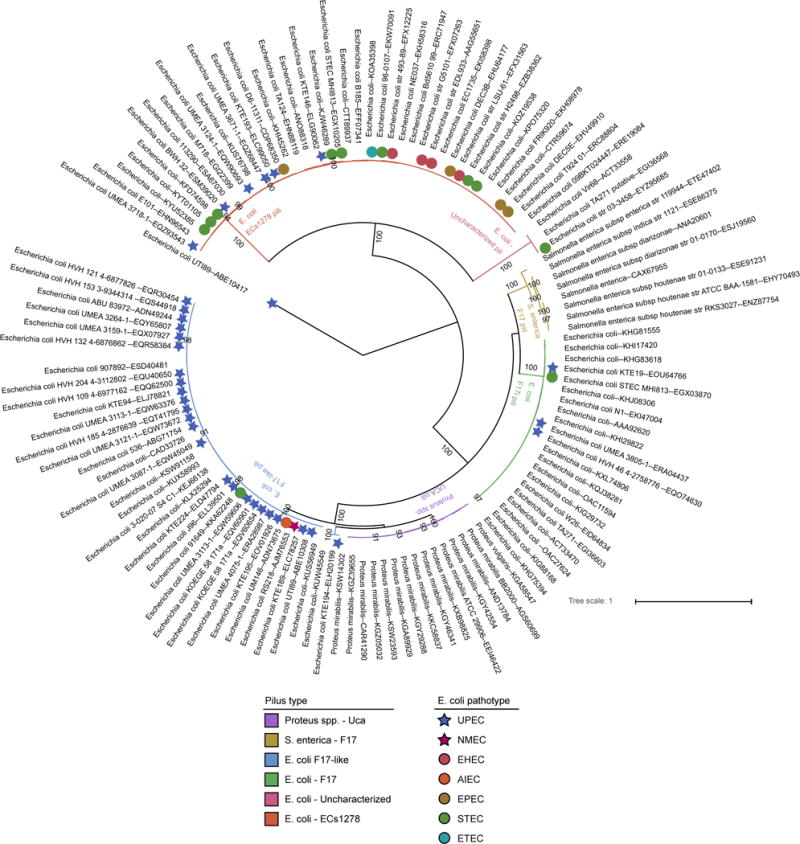
The phylogenetic relationships between F17 homologs was estimated using the sequence of the usher genes. Branch colors indicate host strain and pilus identity, while colored symbols indicate the annotated pathotype of the E. coli strain for each sequence as determined by publically available annotations. Stars indicate extraintestinal pathogenic E. coli (ExPEC) strains while circles indicate intestinal pathogenic E. coli strains. Carriage of F17-like pili is enriched in UPEC strains while F17 and ECs1278 pili are more common in intestinal pathogens such as EHEC. The strain names for each sequence and ENA accession IDs are given. Numbers beneath the branches indicate the percentage of support from 1000 bootstrap replicates (numbers greater than 80% are shown).
Extended Data Figure 5. Phylogenetic distribution of F17-like carriage in UPEC from patients with rUTI.
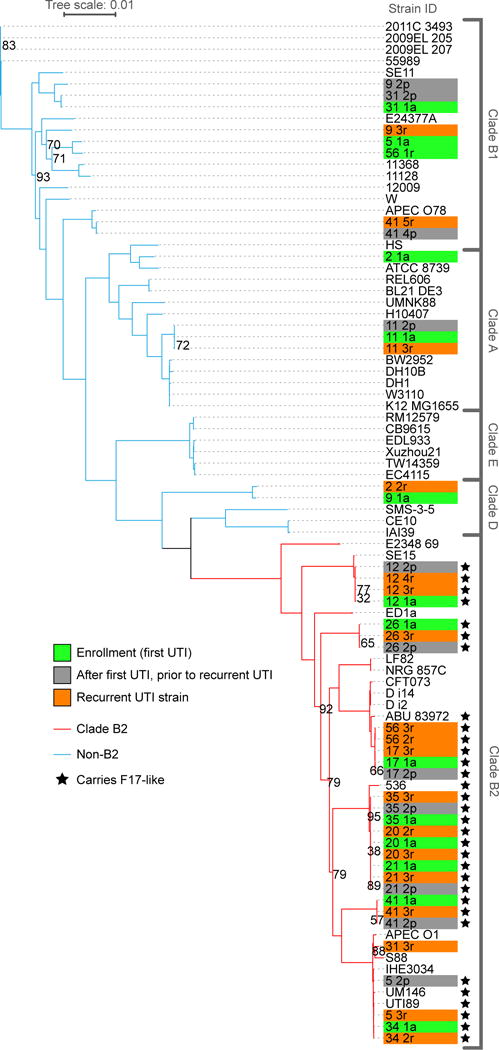
The phylogeny of a set of clinical UPEC strains (n=43 with taxon labels highlighted in green, orange, or grey) was contextualized with reference E. coli strains (n=46, unhighlighted taxon labels) by comparing the concatenated single-copy, core genes of the strains using the RAxML algorithm and the GTRCAT model29. Highlighted taxon labels indicate UPEC isolates collected at enrollment (green) and during recurrent UTI (orange). In all cases, patients cleared each infection prior to recurrence, no patient exhibited signs of asymptomatic bacteriuria. The study design also allowed for the collection, from cohort participants, of E. coli isolates present in the urine in the days leading up to their clinical visit and rUTI diagnosis (highlighted in grey)17. Branch lines indicate phylogenetic background for strains from clade B2 (red branch lines) and non-B2 clades (blue branch lines). Carriage of F17-like pili (black stars) was limited to the B2 clade and enriched within rUTI UPEC isolates. Bootstrap supports are indicated at internal nodes. Bootstrap values >95 have been removed. The clade to which each strain belongs is indicated in brackets to the right.
Extended Data Figure 6. Testing the effects of more prolonged dosing of M4284 and analysis of the duration of its effects.
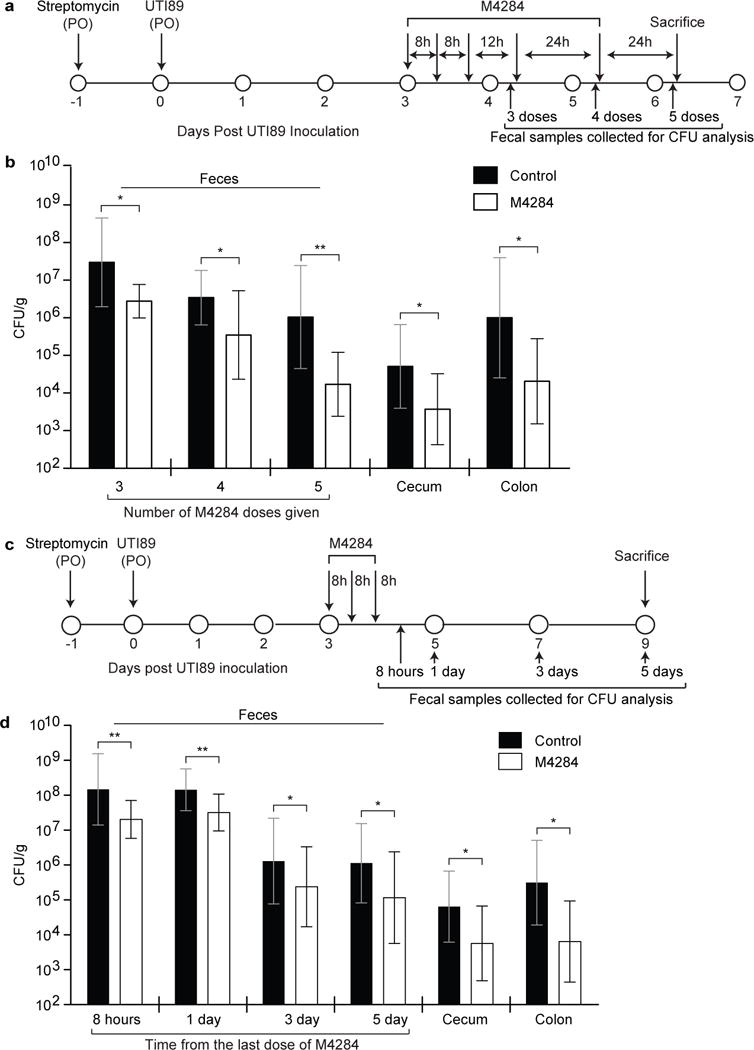
(a) Experimental design. (b) Animals treated as in panel (a) show a continued decrease in UTI89 levels in their feces (samples were processed after 3, 4, and 5 doses of M4284), and at the time of sacrifice in the cecum and colon, compared to control mice treated with vehicle alone (Control, 10% cyclodextrin). (c,d) The effects of mannoside treatment persist 5 days after M4284 exposure. Bars represent geometric mean ± SD, *p<0.05, **p<0.01 by Mann Whitney U test. n=9 mice (Control); n=10 mice (M4284), 2 biological replicates (b). n=16 mice, 3 biological replicates (d).
Extended Data Figure 7. The severity of UTI outcome is directly linked to the dose of UTI89 inoculated into the urinary tract.
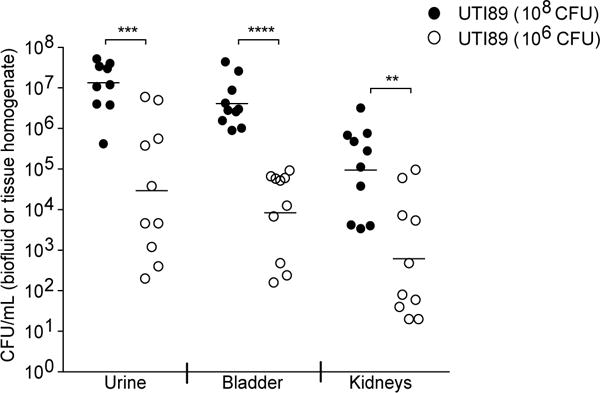
C3H/HeN mice (Envigo) were given an experimental UTI via transurethral introduction of either 106 or 108 CFU of UTI89. The doses were chosen to represent the reduction observed in intestinal UTI89 titers before and after treatment with the M4284 mannoside. Mice were sacrificed 24 hours after inoculation and UTI89 titers in urine, bladder, and kidneys were defined by quantifying CFU. Mice receiving the 106 dose of UTI89 had significantly fewer bacteria in all three biospecimen types, indicating an important relationship between the number of bacteria introduced into the urinary tract and the severity of UTI outcome. Bars represent geometric means, **p<0.01, ***p<0.001, ****p<0.0001 by Mann Whitney U test. n=10 mice, 2 biological replicates.
Extended Data Figure 8. 16S rRNA-based comparison of fecal bacterial communities in mice obtained from Envigo and Charles Rivers Labs and mice of different genetic backgrounds from a common vendor.
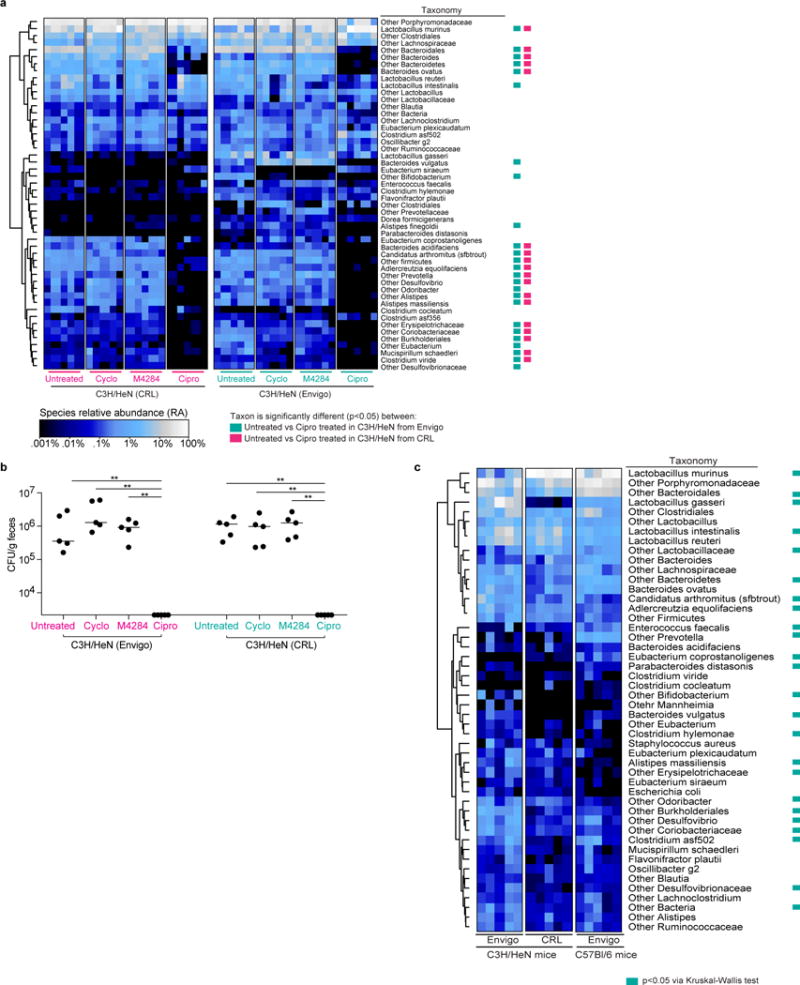
(a) C3H/HeN mice were treated with M4284 (100mg/kg, 3 doses over 24 h), vehicle alone (10% cyclodextrin (Cyclo), 3 doses over 24 h), or ciprofloxacin (Cipro; 15mg/kg, 2 doses over 24 h). Untreated mice served as reference controls. Heatmaps show the effect of each of the treatments on animals from CRL and Envigo. Each row represents a species-level bacterial taxon, while each column represents a mouse sampled 24 hours after the termination of the indicated treatment. Colored boxes next to the taxon names indicate species whose relative abundance was significantly changed by Cipro treatment (p<0.05;Wilcoxon Signed Rank test with FDR correction). Individual comparisons between untreated and other treatment types did not disclose changes that were statistically significant by Wilcoxon Signed Rank test with FDR correction. (b) Corresponding fecal samples collected 24 h after treatments (as shown in Extended Data Figure 8a) were homogenized, diluted serially, and plated on MacConkey medium. The abundance of bacteria capable of growing on the selective medium was similar between fecal samples taken from untreated mice and those collected 24 hours after treatment with cyclodextrin and M4284. No colonies were detected from fecal samples collected 24 hours after ciprofloxacin treatment. (c) Comparison of the representation of bacterial taxa in the fecal microbiota of untreated mice obtained from different vendors or representing different genetic backgrounds. Each row in the heatmap represents a species-level taxon, while each column represents a mouse of the indicated genetic background from the indicated vendor. Colored boxes indicate species whose relative abundances were significantly different (p<0.05) between all three groups of animals (Kruskal-Wallis test with FDR correction). Rows of each heatmap were hierarchically clustered according to pair-wise distances using Pearson correlation. n=5 mice per treatment type, 1 biological replicate (a). n=5 mice, 1 biological replicate (b). n= 5 mice per vendor/mouse strain, 1 biological replicate (c). bar = median; **p<0.001, Mann Whitney U test (b).
Extended Data Figure 9. The configuration of the fecal microbiota of C3H/HeN mice pretreated with streptomycin and colonized with UTI89 is minimally altered by M4284 treatment.
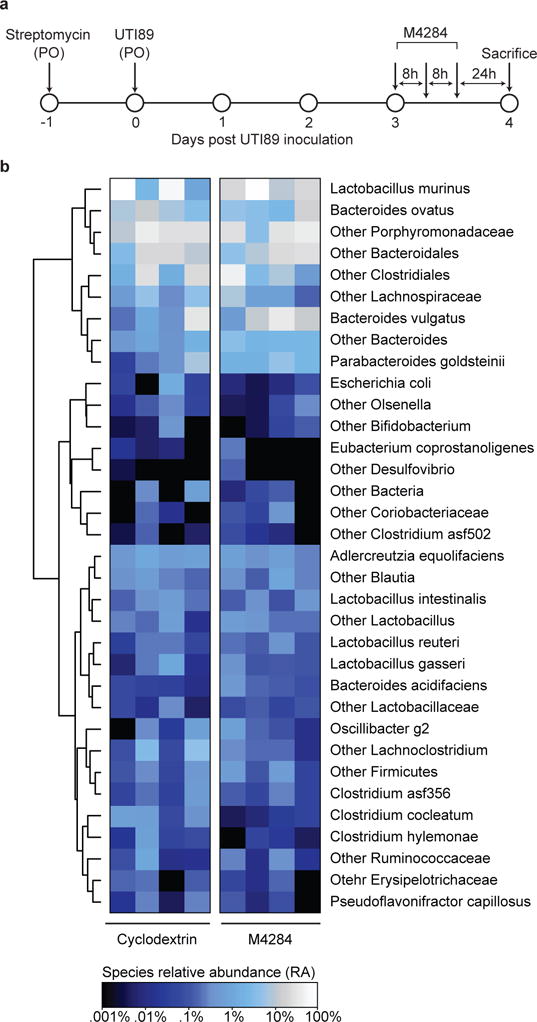
(a) C3H/HeN mice from Envigo were pretreated with streptomycin and 24 h later colonized with UTI89 by oral gavage. Three days after inoculation, animals were treated with 3 doses of M4284 (100mg/kg, 3 doses over 24 h) or vehicle alone (10% cyclodextrin (Cyclo), 3 doses over 24 h). Fecal samples were collected 24 h after the last dose of M4284 or vehicle. (b) Heatmap showing the effect of each treatment type. Each row represents a bacterial species-level taxon, while each column represents a mouse 24 h after the indicated treatment. Rows of the heatmap were hierarchically clustered according to pair-wise distances using Pearson correlation. No treatments produced changes that were statistically significant, as judged by Wilcoxon Signed Rank test with FDR correction. n=4 mice per treatment type, 1 biological replicate.
Extended Data Figure 10. The percent reduction in strains by M4284 treatment is similar in mice colonized with genetically distinct human isolates, and in multiple strains of mice colonized with UTI89.
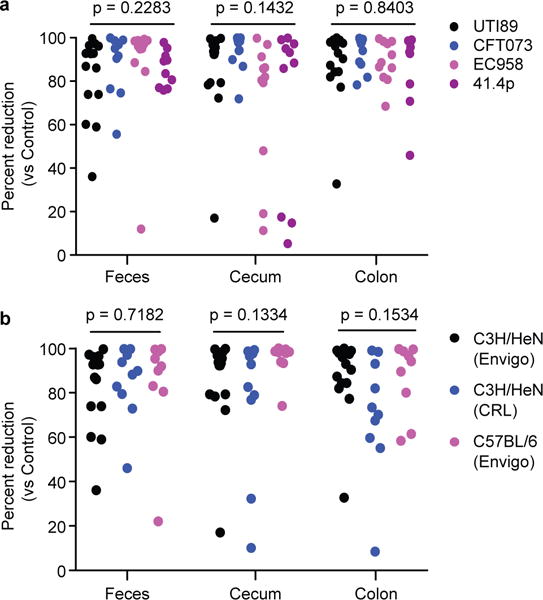
(a) The percent reduction in CFU for the indicated UPEC strains from M4284-treated versus untreated control C3H/HeN mice obtained from Envigo (based on data presented in Fig. 4c–f). (b) CFU data obtained from CRL C3H/HeN mice and Envigo C57BL/6 mice (based on data shown in Fig. 4f–h). P values calculated using Kruskal-Wallis test. n=14 mice, 3 biological replicates (UTI89), n=10 mice, 2 biological replicates (CFT073, EC958, 41.4p) (a). n=14 mice, 3 biological replicates (C3H/HeN from Envigo), n=10 mice, 2 biological replicates (C3H/HeN from CRL), and n=9 mice, 2 biological replicates (C57BL/6 from Envigo) (b).
Supplementary Material
Acknowledgments
Ashlee Earl, Abigail Manson for assistance in acquiring genomic data, Jay Nix and Molecular Biology Consortium (Beamline 4.2.2) for collection/processing crystallographic data, Lars Vereecke and Amanda Goncalves at VIB Bio Imaging Core for support with tissue sectioning/microscopy, Zhenfu Han for synthesizing M4284, Kevin Tamadonfar for assistance with tissue binding studies, and Magdalena Lukaszczyk with protein production. This work was supported by grants from the NIH [K08AI113184 (ALK), R01AI048689 (SJH), P50DK064540 (SJH), RC1DK086378 (SJH), DK30292 (JIG) and 1F31DK107057 (CNS)], FWO-Flanders (G030411N), Hercules Foundation (UABR/09/005) and VIB PRJ9.
Footnotes
Author Contributions: SJH, JIG, CNS developed the mouse model and identified relevant CUP pili. SJH, JIG, CNS, JWJ, ZTC designed and CNS performed M4284 experiments. JWJ provided M4284. SJH, JIG, ALK, CNS executed microbiota experiments. HR, SR, RDK, CNS designed and RDK, SR performed in vitro binding assays. JSP, KWD expressed and purified protein. SR, HR, DHF, RDK, KWD performed crystallization trials and solved UclD structures. HR, SJH, HLS, CNS designed and HLS performed phylogenetic and genomic analyses. SJH, JIG, HR, CNS wrote the manuscript with input from all authors.
Competing Financial Interests (CFI): JWJ and SJH are inventors on patent application US8937167, which covers the use of mannoside-based FimH ligand antagonists for the treatment of disease. JWJ and SJH have ownership interest in Fimbrion Therapeutics, and may benefit if the company is successful in marketing mannosides.
References
- 1.Foxman B. Urinary tract infection syndromes: occurrence, recurrence, bacteriology, risk factors, and disease burden. Infect Dis Clin North Am. 2014;28:1–13. doi: 10.1016/j.idc.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 2.Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nature Rev Microbiol. 2015;13:269–284. doi: 10.1038/nrmicro3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zowawi HM, et al. The emerging threat of multidrug-resistant Gram-negative bacteria in urology. Nature Reviews Urology. 2015;12:570–584. doi: 10.1038/nrurol.2015.199. [DOI] [PubMed] [Google Scholar]
- 4.Mediavilla JR, et al. Colistin- and Carbapenem-Resistant Escherichia coli Harboring mcr-1 and blaNDM-5, Causing a Complicated Urinary Tract Infection in a Patient from the United States. mBio. 2016;7:e01191–16. doi: 10.1128/mBio.01191-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spaulding CN, Hultgren SJ. Adhesive Pili in UTI Pathogenesis and Drug Development. Pathogens. 2016;5:e30. doi: 10.3390/pathogens5010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamamoto S, et al. Genetic evidence supporting the fecal-perineal-urethral hypothesis in cystitis caused by Escherichia coli. J Urology. 1997;157:1127–1129. [PubMed] [Google Scholar]
- 7.Wurpel DJ, Beatson SA, Totsika M, Petty NK, Schembri MA. Chaperone-usher fimbriae of Escherichia coli. PloS one. 2013;8:e52835. doi: 10.1371/journal.pone.0052835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaiser P, Diard M, Stecher B, Hardt WD. The streptomycin mouse model for Salmonella diarrhea: functional analysis of the microbiota, the pathogen’s virulence factors, and the host’s mucosal immune response. Immunol Rev. 2012;245:56–83. doi: 10.1111/j.1600-065X.2011.01070.x. [DOI] [PubMed] [Google Scholar]
- 9.Wright KJ, Seed PC, Hultgren SJ. Development of intracellular bacterial communities of uropathogenic Escherichia coli depends on type 1 pili. Cellular Microbiol. 2007;9:2230–2241. doi: 10.1111/j.1462-5822.2007.00952.x. [DOI] [PubMed] [Google Scholar]
- 10.Wurpel DJ, et al. Comparative proteomics of uropathogenic Escherichia coli during growth in human urine identify UCA-like (UCL) fimbriae as an adherence factor involved in biofilm formation and binding to uroepithelial cells. J Proteomics. 2016;131:177–189. doi: 10.1016/j.jprot.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 11.Jarvis C, et al. Antivirulence Isoquinolone Mannosides: Optimization of the Biaryl Aglycone for FimH Lectin Binding Affinity and Efficacy in the Treatment of Chronic UTI. Chem Med Chem. 2016;11:367–373. doi: 10.1002/cmdc.201600006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nuccio S, Baumler AJ. Evolution of the chaperone/usher assembly pathway: fimbrial classification goes Greek. Microbiol Mol Biol Rev. 2007;71:551–575. doi: 10.1128/MMBR.00014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang L, Foxman B, Marrs C. Both urinary and rectal Escherichia coli isolates are dominated by strains of phylogenetic group B2. J Clin Microbiol. 2002;40:3951–3955. doi: 10.1128/JCM.40.11.3951-3955.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richards VP, et al. Genome based phylogeny and comparative genomic analysis of intra-mammary pathogenic Escherichia coli. PloS one. 2015;10:e0119799. doi: 10.1371/journal.pone.0119799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Low AS, et al. Analysis of fimbrial gene clusters and their expression in enterohaemorrhagic Escherichia coli O157:H7. Environ Microbiol. 2006;8:1033–1047. doi: 10.1111/j.1462-2920.2006.00995.x. [DOI] [PubMed] [Google Scholar]
- 16.Merckel MC, et al. The structural basis of receptor-binding by Escherichia coli associated with diarrhea and septicemia. J Mol Biol. 2003;331:897–905. doi: 10.1016/s0022-2836(03)00841-6. [DOI] [PubMed] [Google Scholar]
- 17.Czaja CA, et al. Prospective cohort study of microbial and inflammatory events immediately preceding Escherichia coli recurrent urinary tract infection in women. J Infectious Dis. 2009;200:528–536. doi: 10.1086/600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schreiber HL, et al. Bacterial virulence phenotypes of Escherichia coli and host susceptibility determine risk for urinary tract infections. Science Translational Med. 2017;9:eaaf1283. doi: 10.1126/scitranslmed.aaf1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han Z, et al. Lead optimization studies on FimH antagonists: discovery of potent and orally bioavailable ortho-substituted biphenyl mannosides. J Med Chem. 2012;55:3945–3959. doi: 10.1021/jm300165m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cusumano CK, et al. Treatment and prevention of urinary tract infection with orally active FimH inhibitors. Science Translational Med. 2011;3:109–115. doi: 10.1126/scitranslmed.3003021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosen DA, Hung CS, Kline KA, Hultgren SJ. Streptozocin-induced diabetic mouse model of urinary tract infection. Infection Immun. 2008;76:4290–4298. doi: 10.1128/IAI.00255-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones CH, et al. FimH adhesin of type 1 pili is assembled into a fibrillar tip structure in the Enterobacteriaceae. Proc Natl Acad Sci USA. 1995;92:2081–2085. doi: 10.1073/pnas.92.6.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Totsika M, et al. Insights into a multidrug resistant Escherichia coli pathogen of the globally disseminated ST131 lineage: genome analysis and virulence mechanisms. PloS one. 2011;6:e26578. doi: 10.1371/journal.pone.0026578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Welch RA, et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc Natl Acad Sci USA. 2002;99:17020–17024. doi: 10.1073/pnas.252529799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yao J, et al. A Pathogen-Selective Antibiotic Minimizes Disturbance to the Microbiome. Antimicrobial Agents and Chemotherapy. 2016 doi: 10.1128/AAC.00535-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galtier M, et al. Bacteriophages to reduce gut carriage of antibiotic resistant uropathogens with low impact on microbiota composition. Environ Microbiol. 2016;7:1462–2920. doi: 10.1111/1462-2920.13284. [DOI] [PubMed] [Google Scholar]
- 27.Cox LM, et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell. 2014;158:705–721. doi: 10.1016/j.cell.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wright KJ, Seed PC, Hultgren SJ. Development of intracellular bacterial communities of uropathogenic Escherichia coli depends on type 1 pili. Cellul Microbiol. 2007;9:2230–2241. doi: 10.1111/j.1462-5822.2007.00952. [DOI] [PubMed] [Google Scholar]
- 32.Hung CS, Dodson KW, Hultgren SJ. A murine model of urinary tract infection. Nature Protocols. 2009;4:1230–1243. doi: 10.1038/nprot.2009.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Justice SS, Lauer SR, Hultgren SJ, Hunstad DA. Maturation of intracellular Escherichia coli communities requires SurA. Infect and Immun. 2006;74:4793–4800. doi: 10.1128/IAI.00355-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johansson ME, Hansson GC. Preservation of mucus in histological sections, immunostaining of mucins in fixed tissue, and localization of bacteria with FISH. Methods Molec Biol. 2012;842:229–235. doi: 10.1007/978-1-61779-513-8_13. [DOI] [PubMed] [Google Scholar]
- 35.Kearse M, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Molec Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 38.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kalas V, et al. Evolutionary fine-tuning of conformational ensembles in FimH during host-pathogen interactions. Sci Adv. 2017;3:e1601944. doi: 10.1126/sciadv.1601944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caporaso JG, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rideout JR, et al. Subsampled open-reference clustering creates consistent, comprehensive OTU definitions and scales to billions of sequences. PeerJ. 2014;2:e545. doi: 10.7717/peerj.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kau AL, et al. Functional characterization of IgA-targeted bacterial taxa from undernourished Malawian children that produce diet-dependent enteropathy. Science Translational Med. 2015;7:276ra224. doi: 10.1126/scitranslmed.aaa4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kabsch W. Xds. Acta Crystallographica Section D, Biological Crystallography. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallographica Section D, Biological Crystallography. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D, Biological Crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Bacterial V4-16S rRNA datasets have been deposited in the European Nucleotide Archive (ENA) under accession number PRJEB19121. Sequences used to examine the carriage of F17-like pili in clinical rUTI isolates were previously published18 and are deposited in the NCBI under the Bioproject ID: PRJNA269984. Crystallography data have been deposited in the PDB under accession codes: 5NWP (P21) and 5VQ5 (P212121).


