Extended Data Figure 5. Phylogenetic distribution of F17-like carriage in UPEC from patients with rUTI.
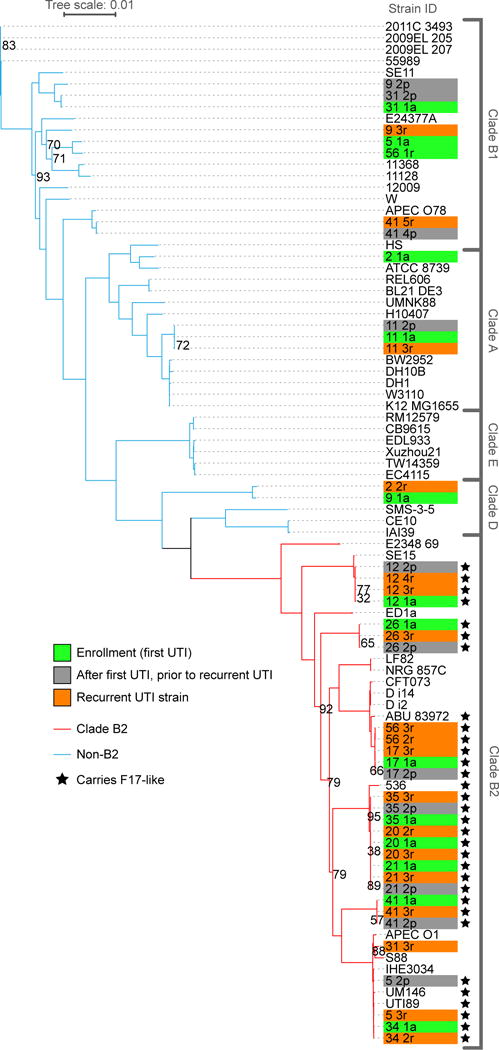
The phylogeny of a set of clinical UPEC strains (n=43 with taxon labels highlighted in green, orange, or grey) was contextualized with reference E. coli strains (n=46, unhighlighted taxon labels) by comparing the concatenated single-copy, core genes of the strains using the RAxML algorithm and the GTRCAT model29. Highlighted taxon labels indicate UPEC isolates collected at enrollment (green) and during recurrent UTI (orange). In all cases, patients cleared each infection prior to recurrence, no patient exhibited signs of asymptomatic bacteriuria. The study design also allowed for the collection, from cohort participants, of E. coli isolates present in the urine in the days leading up to their clinical visit and rUTI diagnosis (highlighted in grey)17. Branch lines indicate phylogenetic background for strains from clade B2 (red branch lines) and non-B2 clades (blue branch lines). Carriage of F17-like pili (black stars) was limited to the B2 clade and enriched within rUTI UPEC isolates. Bootstrap supports are indicated at internal nodes. Bootstrap values >95 have been removed. The clade to which each strain belongs is indicated in brackets to the right.
