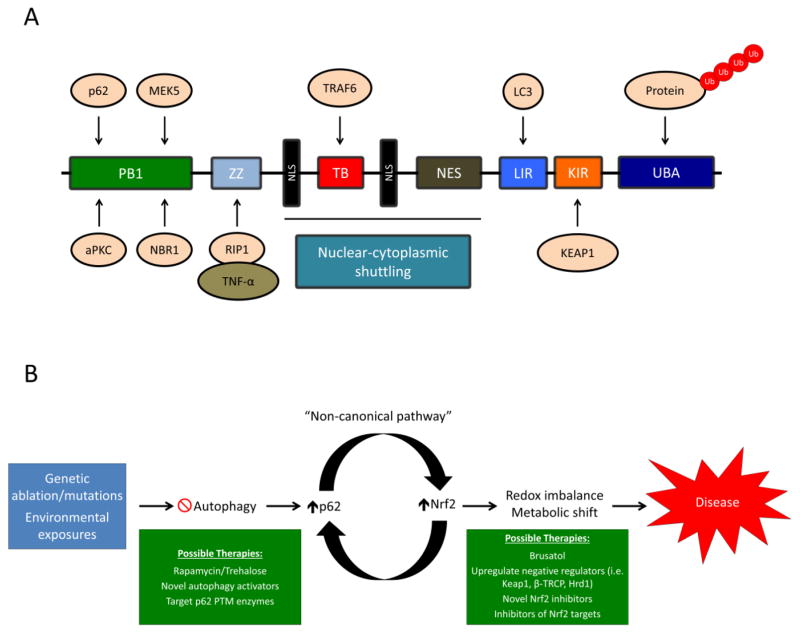
Figure 1. The domain structure of p62 and possible therapeutic interventions to treat the p62-dependent activation of NRF2.
(A) p62 is a multifunctional protein consisting of the following domains: 1) PB1 domain, responsible for p62 self-oligomerization and binding to other PB1 containing proteins (i.e. aPKC, MEK5, and NBR1, shown above); 2) ZZ domain, interacts with RIP-1, which recruits TNF-α and mediates cell death; 3) TB domain, binds to TRAF6, which mediates protein ubiquitylation; 4–6) NLS and NES domains, control the nuclear-cytoplasmic shuttling of p62; 7) LIR, binds to LC3 in the autophagosomal membrane, recruiting cargo for autophagic degradation; 8) KIR, binds to KEAP1; and 9) UBA domain, binds to poly-ubiquitylated proteins. (B) Possible autophagy and NRF2-based therapeutic interventions for treating diseases associated with non-canonical activation of NRF2. Genetic alterations to key autophagy proteins, or chronic exposure to environmental toxicants (i.e. arsenic), result in autophagy blockage and the p62-dependent upregulation of NRF2. Prolonged activation of NRF2 results in changes to the cellular metabolic and redox state, contributing to the pathogenesis of certain diseases. Restoring autophagic function via autophagy activating compounds, preventing the post-translational modifications of p62, or directly inhibiting the function of NRF2 or its downstream targets, could prove viable therapeutic options in the treatment of diseases associated with increased levels of p62 and NRF2.