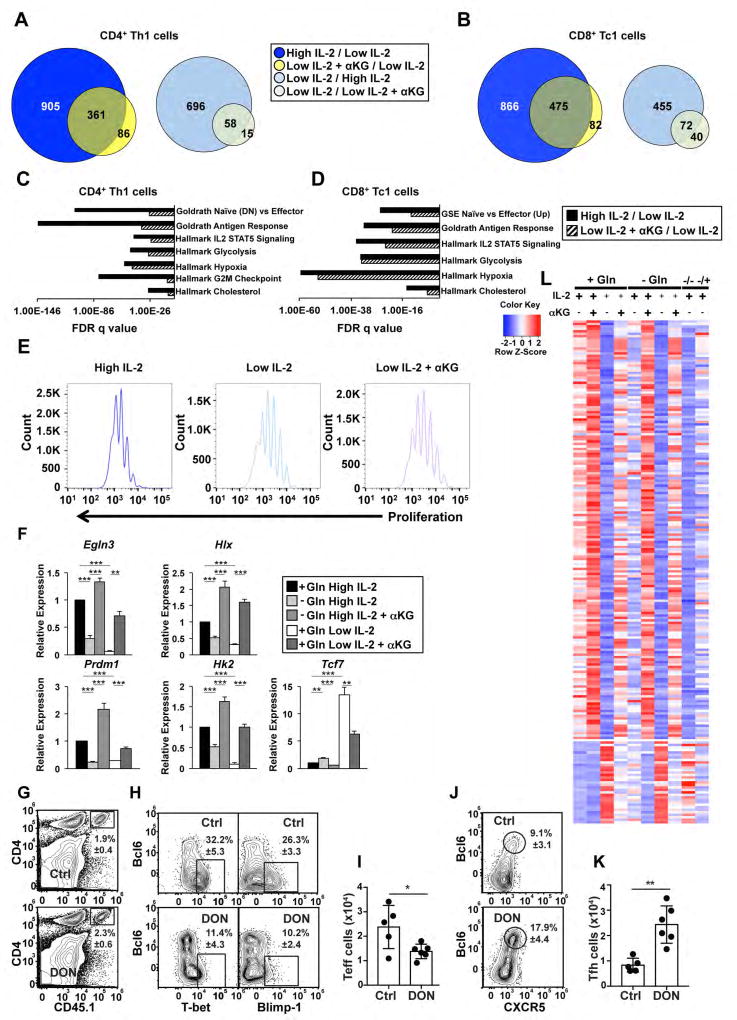
Figure 2. Gln- and α KG-sensitive events regulate a portion of the IL-2-sensitive pathway.
(A, B) Venn diagrams comparing genes that were induced (left) or inhibited (right) by at least 2-fold in high IL-2 (blue circles) or the addition of αKG to low IL-2 (yellow circles) as compared to low IL-2 conditions in (A) CD4+ or (B) CD8+ T cells. (C, D) FDR q values for the enrichment of select pathways from a GSEA examining genes from (C) CD4+ or (D) CD8+ T cells that were induced 2-fold in high IL-2 relative to low IL-2 (black bars) or low IL-2 with αKG relative to low IL-2 (hatched bars). (E) Flow cytometry analysis of E450 dilution in primary CD4+ T cells polarized in Th1 conditions and exposed to high IL-2 (dark blue), low IL-2 (light blue), or low IL-2 with αKG (purple) for two days. Data are representative of 3 independent biological replicates. (F) qRT-PCR analysis of transcript expression from primary CD4+ T cells polarized in Th1 conditions that were maintained in media with Gln and in high IL-2 conditions (black bar), low IL-2 (white bar), or low IL-2 with αKG (dark grey bar) or were maintained without Gln and in high IL-2 (light grey) or high IL-2 with αKG (medium grey) for two days. The n= at least 4 independent biological replicates for each gene. Error bars represent SEM and an unpaired student t-test was performed with p values indicated (* ≤0.05, **≤0.001, and ***≤0.0001). (G) The frequency of CD45.1+OTII cells in the recipients of control (Ctrl) and DON treated cells. (H, J) The frequency and (I, K) number of CD45.1+OTII cells with a (H, I) Teff phenotype (Bcl6loCXCR5loTbethiBlimp1hi) or (J, K) Tfh cell phenotype (Bcl6hiCXCR5hiTbetloBlimp1lo) in recipients receiving control or DON treated cells. Data are shown as the mean ± SD (n=5–6 mice) and are representative of two independent experiments. P values were determined using a two-tailed Student´s t-test (* ≤0.05, **≤0.005). (L) Heatmap displaying Z-scores for genes that were induced (top) or inhibited (bottom) by at least 2-fold in comparison to cells maintained without Gln. Conditions are shown above each lane (see STAR methods for detailed description of design). See also Fig. S1–3.