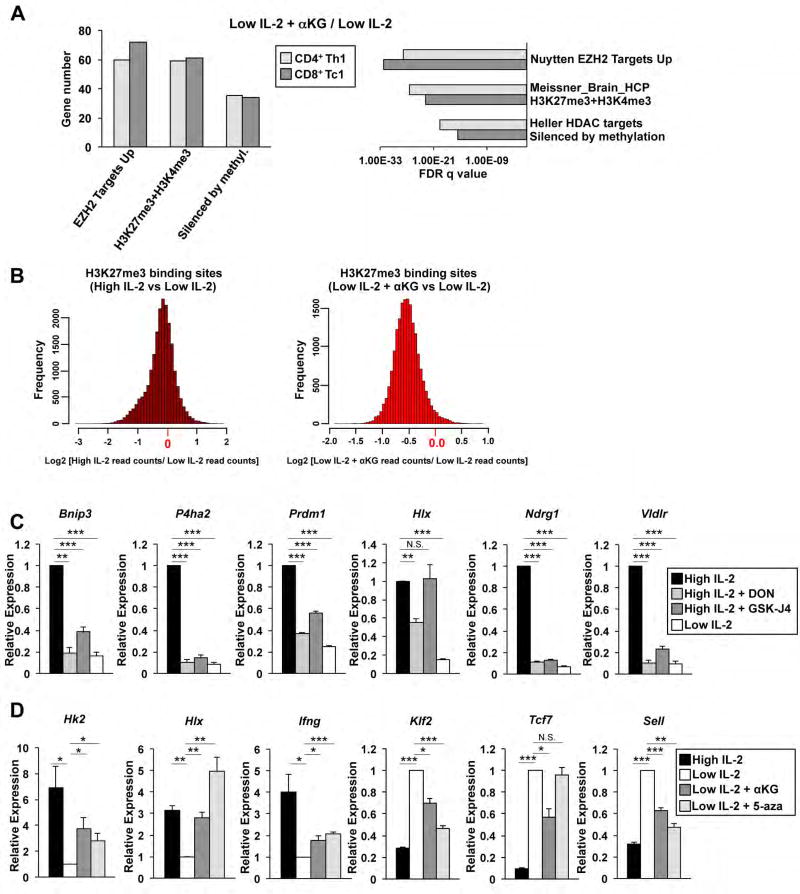
Figure 3. IL-2 and αKG-sensitive events are related to H3K27me3 and DNA methylation states.
(A) GSEA of select chemical and genetic perturbations datasets examining genes induced in the low IL-2 with αKG compared to the low IL-2 condition in CD4+ Th1 (light grey) or CD8+ Tc1 (dark grey) cells. (B) Histograms representing differential H3K27me3 ChIP-seq peak read depth enrichment between high IL-2 compared to low IL-2 conditions (left) or low IL-2 with αKG relative to low IL-2 conditions (right). The distribution of the log ratio of peak read enrichment is displayed. (C, D) qRT-PCR analysis of transcripts from primary CD4+ Th1 cells exposed to (C) high IL-2 (black bars), low IL-2 (white bars), high IL-2 with DON (light grey bars) or high IL-2 with GSK-J4 (dark grey bars) or (D) high IL-2 (black bars), low IL-2 (white bars), low IL-2 with αKG (dark grey bars), or low IL-2 with 5-azacytidine (light grey bars). (C, D) The n= at least (C) 4 except high IL-2 with DON n= 3 or (D) 3 independent biological replicates for each gene. Error bars represent SEM and an unpaired student t-test was performed with p values indicated (* ≤0.05, **≤0.001, and ***≤0.0001). See also Fig. S3.