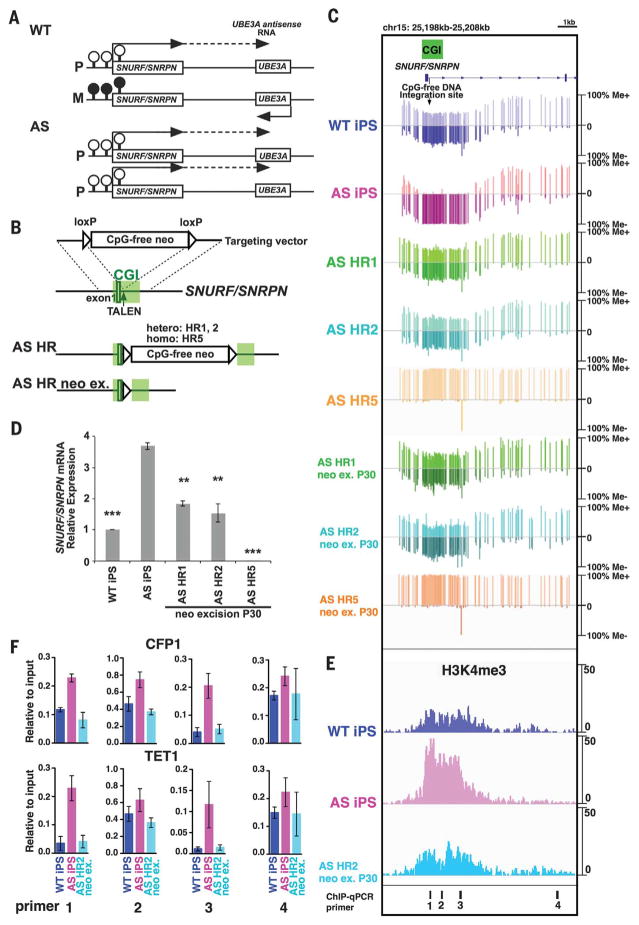
Fig. 2. Targeted integration of the CpG-free cassette corrects the aberrant DNA methylation in AS iPSCs.
(A) Schematic of allele-specific methylation status and gene expression on the imprinting locus chr15q11-q12 in neurons of a normal individual and an AS patient with paternal UPD. P and M represent paternal and maternal allele, respectively. Filled and open circles represent methylated and unmethylated CpGs on the CGI, respectively. The dashed line represents a long noncoding RNA. (B) Schematic of HR with CpG-free donor vector targeting the SNURF/SNRPN gene locus. TALEN, transcription activator–like effector nuclease. (C) Target enrichment–genome bisulfite sequencing analysis showing methylation status of SNURF/SNRPN locus in iPSCs. See also fig. S10, showing the whole region. (D) Quantitative RT-PCR analysis of SNURF/SNRPN mRNA expression normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA in iPSCs. Error bars indicate ±SEM of three independent experiments. Statistical significance was determined by one-way analysis of variance (ANOVA) followed by Bonferroni’s multiple-comparison test (versus AS iPSCs; **P < 0.001; ***P < 0.0001). (E) Chromatin immunoprecipitation (ChIP)–sequencing data for H3K4me3 across SNURF/SNRPN locus in iPSCs. (F) ChIP–quantitative PCR (qPCR) analysis of CFP1 and TET1 in iPSCs using primer sets targeting de novo–methylated locus inside (primer sets 1 to 3) or outside (primer set 4) the CGI shown in (E). Error bars indicate ±SEM from triplicate experiments.