Fig. 1.
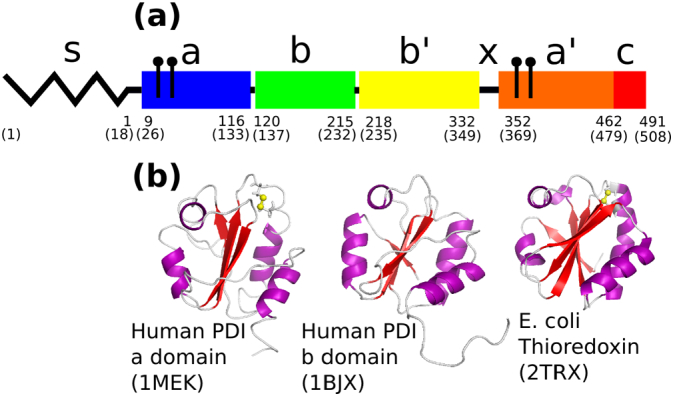
Domain architecture of PDI.
a) Overall domain architecture of human PDI inferred from sequence homology, protease susceptibility and the properties of recombinant constructs representing putative domains [4], [5], [18], [26], [52]. The zig-zag (s) represents the signal sequence that is cleaved during biosynthesis and does not form part of the mature protein. Each coloured block (a, b, b′, a′) represents a domain of the mature protein. Note the original literature uses a residue numbering based on the sequence of mature PDI whereas some later literature uses a residue numbering based on the sequence of the inferred translation product (i.e. including the signal sequence). Residue numbers here and elsewhere in the review are for the mature sequence (above) and for the unprocessed translation product (below, in brackets). ¶ symbols represent cysteine residues of the active sites namely residues 36(53) and 39(56) in the a domain and 380(397) and 383(400) in the a′ domain.
b) Conservation of the thioredoxin-fold tertiary structure in the domains of PDI. The figure shows from left to right in the same orientation, human PDI a domain determined by NMR (PDB id: 1MEK), human PDI b domain determined by NMR (PDB id: 1BJX) and the archetype, E. coli thioredoxin, determined by X-ray crystallography (PDB id: 2TRX). The active site Cys residues in thioredoxin and PDI a domain are shown as ball-and-stick, with the disulfide highlighted in yellow. The PDI b domain does not contain this feature.
